SNP locus primer combination for identifying pinus bungeana germplasm resources and application
A technique of white bark pine and locus, which is applied in the field of identification of white bark pine germplasm resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0148] The preparation method of the above kit includes packaging each primer in any one of the above primer sets separately.
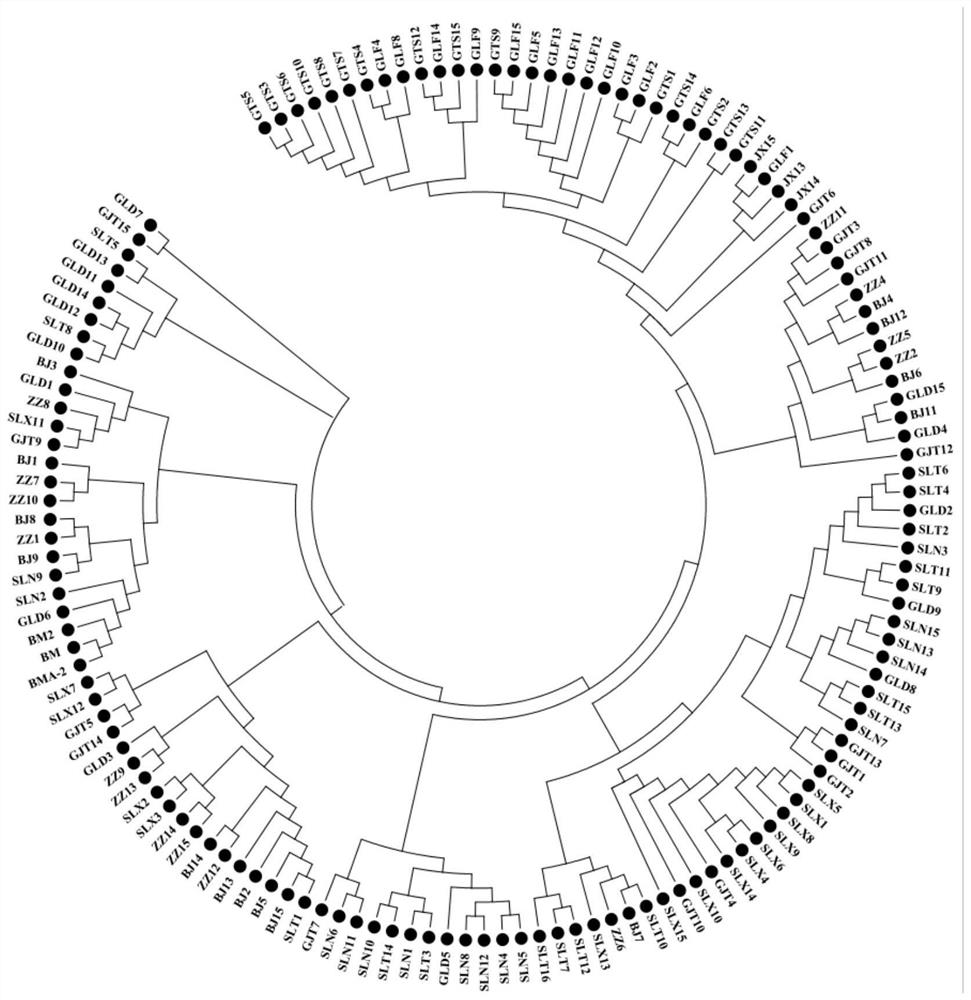
[0149] In the fourth aspect, the present invention provides the above-mentioned DNA fingerprint database of Pine bark germplasm resources based on core SNP markers. The DNA fingerprint database includes: the genotypes of the standard Pine bark germplasm resources at the above 36 SNP sites.
[0150] The above-mentioned standard pine germplasm resources are selected from the following 137 pine germplasm resources:
[0151] Jingxi No. 1, Jingxi No. 11, Jingxi No. 12, Jingxi No. 13, Jingxi No. 14, Jingxi No. 15, Jingxi No. 2, Jingxi No. 3, Jingxi No. 4, Jingxi No. 5, Jingxi No. 6, Jingxi No. 7, Jingxi No. 8, Jingxi No. 9, Penxian, Ruiwang, Nine Trees, Ganjing No. 1, Ganjing No. 10, Ganjing No. 11, Ganjing No. 12, Ganjing No. 13, Ganjing No. 14, Ganjing No. 15, Ganjing No. 2, Ganjing No. 3, Ganjing No. 4, Ganjing No. 5, Ganjing No. 6, Ganjing No. 7, Ganji...
Embodiment 1
[0169] Acquisition of Loci and Primer Combinations for Identification of Pine Pine Germplasm Resources
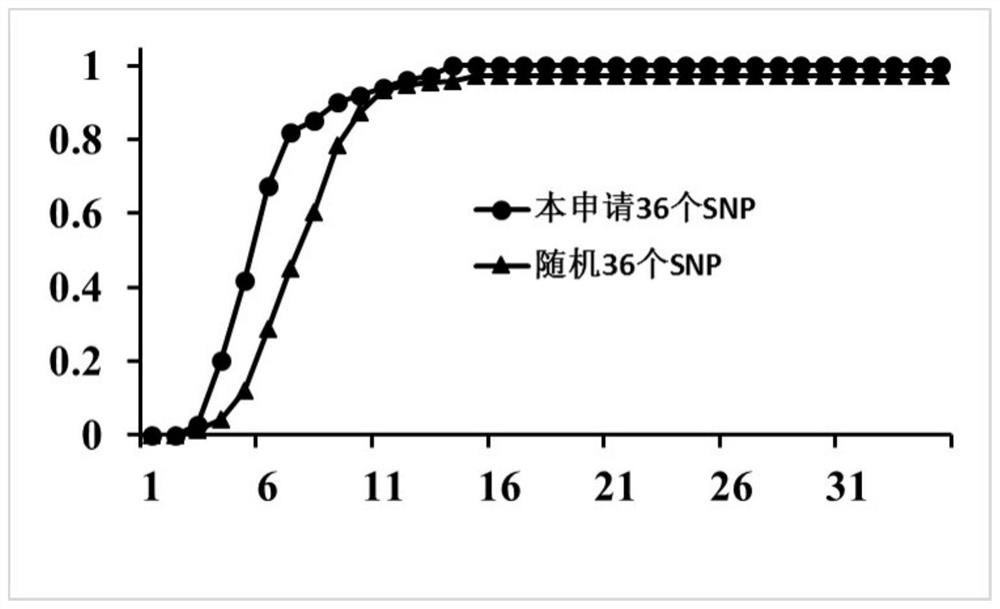
[0170] 1. Discovery of 36 core SNP sites
[0171] The present invention uses an Illumina sequencer, based on the SLAF sequencing (Specific-Locus Amplified Fragment Sequencing) data of 150 pine germplasm resources, and finally obtains 36 SNP sites. These 150 germplasm resources of Pine bark are from different provinces in China, with high genetic diversity and extremely representative genetics.
[0172] SNP site screening criteria are as follows: (1) SLAF sequencing, based on the no-reference genome method, obtained 20,911 SNPs, removed SNP sites with MAF0.3, and heterozygosity rate greater than 0.3, and obtained 7,795 SNPs. (2) Extract the flanking sequence, remove the SNP located within 20bp of the head and tail of the two reads, and intercept 20-40bp from the upstream and downstream according to the actual situation, and obtain 3,843 SNPs. (3) Design Kaspar platform pri...
Embodiment 2
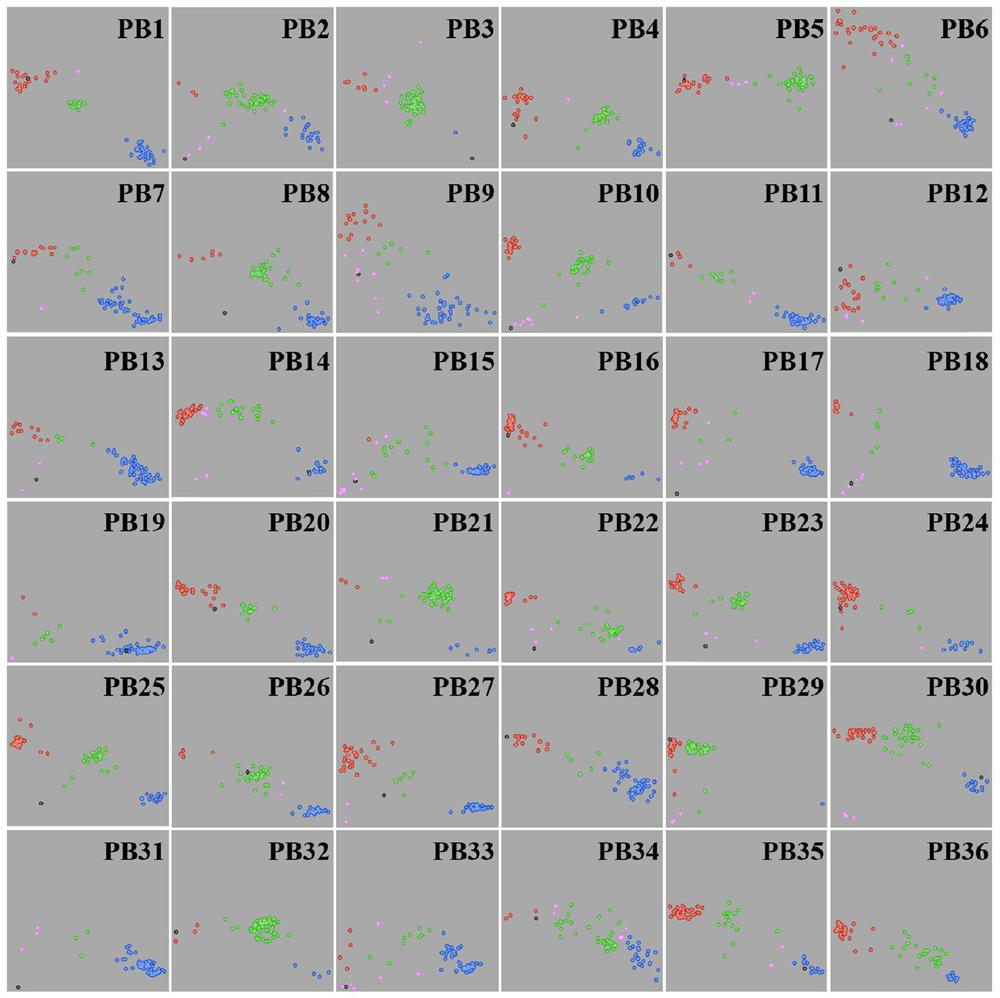
[0190] This example is a test of the effectiveness of the SNP primer combination developed in Example 1, and also the construction of a DNA fingerprint database of Pine bark germplasm resources based on the above-mentioned 36 core SNP markers.
[0191] The 137 pine germplasm resources for testing in the present embodiment are common excellent germplasm resources, and the specific circumstances are as follows in Table 3:
[0192] Table 3: Sources of Germplasm Resources
[0193]
[0194]
[0195] 1. Obtaining the genomic DNA of the tested Pine bark germplasm resources
[0196] Genomic DNA of 137 coniferous needles of pine germplasm resources tested was extracted by CTAB method, and genomic DNA of pine germplasm resources tested was obtained.
[0197] The operation of the above CTAB method is as follows:
[0198] (1) The needles of the above-mentioned 137 pine germplasm resources at the mature stage were harvested, and placed in a freeze dryer (CoolSafe 55-4) for dehydrat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com