DNA methylation markers for noninvasive detection of cancer and uses thereof
A methylation and cancer technology, applied in recombinant DNA technology, DNA/RNA fragments, chemical instruments and methods, etc., can solve problems such as false positives, false negatives, and cell-free DNA.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0307] Example 1. "Classically unmethylated CGIDs found in normal tissue and blood DNA across hundreds of individuals"
[0308]Cell-free DNA derived from tumors is known to be found in body fluids such as plasma, urine and feces. The DNA methylation profile of CF tumor DNA was also determined to be similar to that of tumor DNA (Dominguez-Vigil et al., 2018). A large body of data has established that tumor DNA is differentially methylated compared to normal tissue (Luczak and Jagodzinski, 2006). Consequently, many groups have attempted to use logistic regression to delineate the CGID locations in DNA (CG IDs in the Illumina 450K inventory) that are discriminatively between cancerous tissue and its normal tissue of origin (e.g., liver cancer versus adjacent liver tissue). Basicization. However, since these methods measure quantitative differences between cancer and non-transformed tissue rather than qualitative differences in classification, these quantitative differences betw...
Embodiment 2
[0323] Example 2: "Binary Classification of Differentiation (BCD)" method for the detection of cancer in cell-free DNA.
[0324] The following publicly available database of normalized beta values for methylation of approximately 450,000 CG(ID)s across the human genome was used to derive a list of cancer-specific DNA methylation markers:
[0325] Table 29 Liver cancer
[0326]
[0327]
[0329]
[0330]
[0331] Table 31 Prostate cancer
[0332] disease state source group N Detection / specificity control GSE5295 Find 5 detection PRAD TCGA Find 10 detection PRAD TCGA Find 10 specificity Non-PRAD TCGA Find 80 specificity PRAD GSE7354 verify 77 detection PRAD GSE5295 verify 25 detection PRAD TCGA verify 553 specificity bladder TCGA verify 439 specificity brain TCGA verify 689 specificity breast TCGA verif...
Embodiment 3
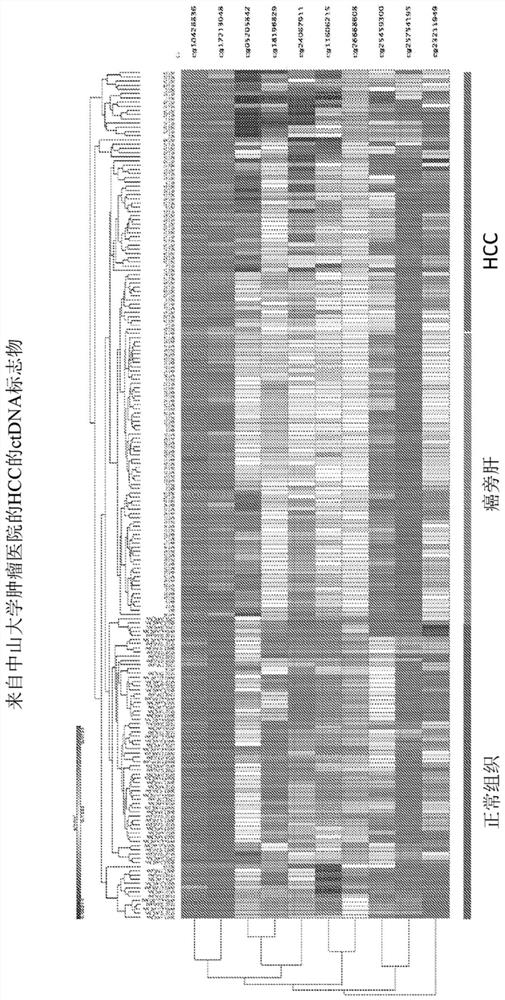
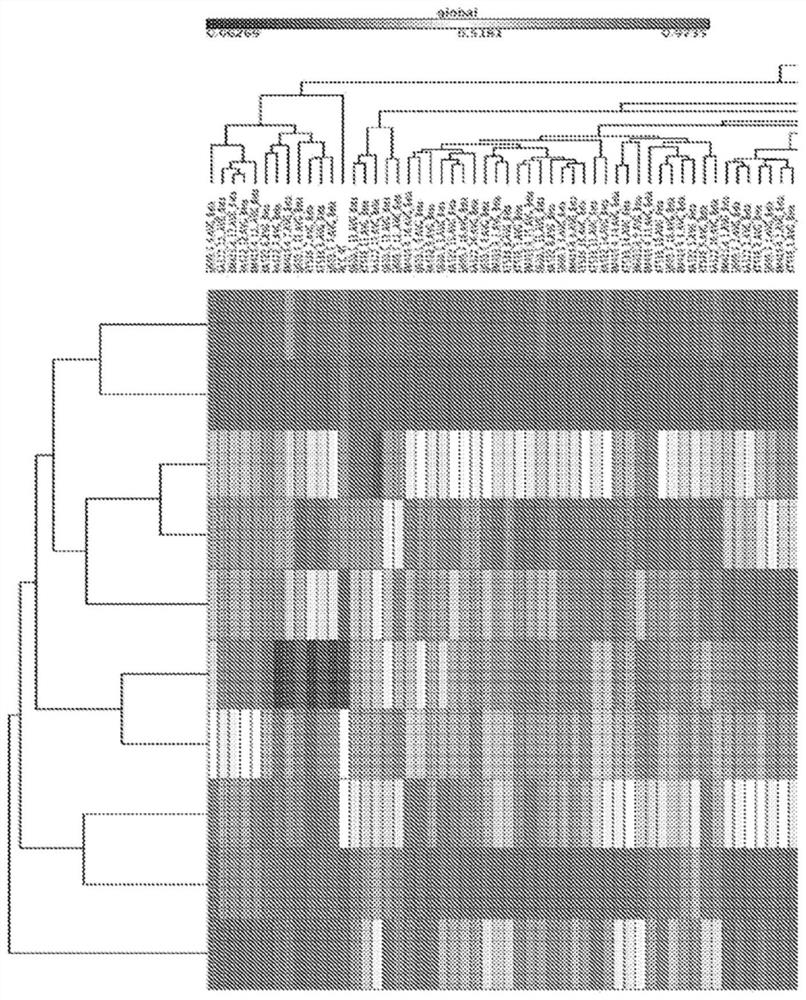
[0398] Example 3. Discovery of multigene DNA methylation markers in liver cancer (HCC).
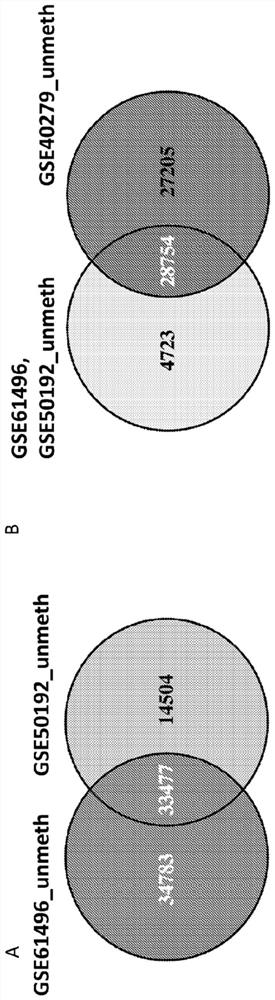
[0399] The inventors used normalized Illumina 450K DNA methylation data from GSE61258 (normal liver) and 66 randomly selected samples from the TCGA HCC collection of HCC DNA methylation data as a "training" cohort. First, the inventors screened the candidate list in the "training cohort" dataset of 28754 CGIDx found in Example 1 to be robustly unmethylated sites across normal tissue and blood samples. The inventors then used the BCD approach described in Example 2 to discover a multi-gene group of binary taxonomically discriminative methylated CGIDs that detected HCC with high sensitivity and specificity in the training cohort ( Figure 5 B, Table 1) (assay). Cancer-specific weighted DNA methylation scores and thresholds were generated for CGID as described in Example 2. The inventors then generated a "training cohort" from 80 randomly selected DNA methylation samples from TCGA represen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com