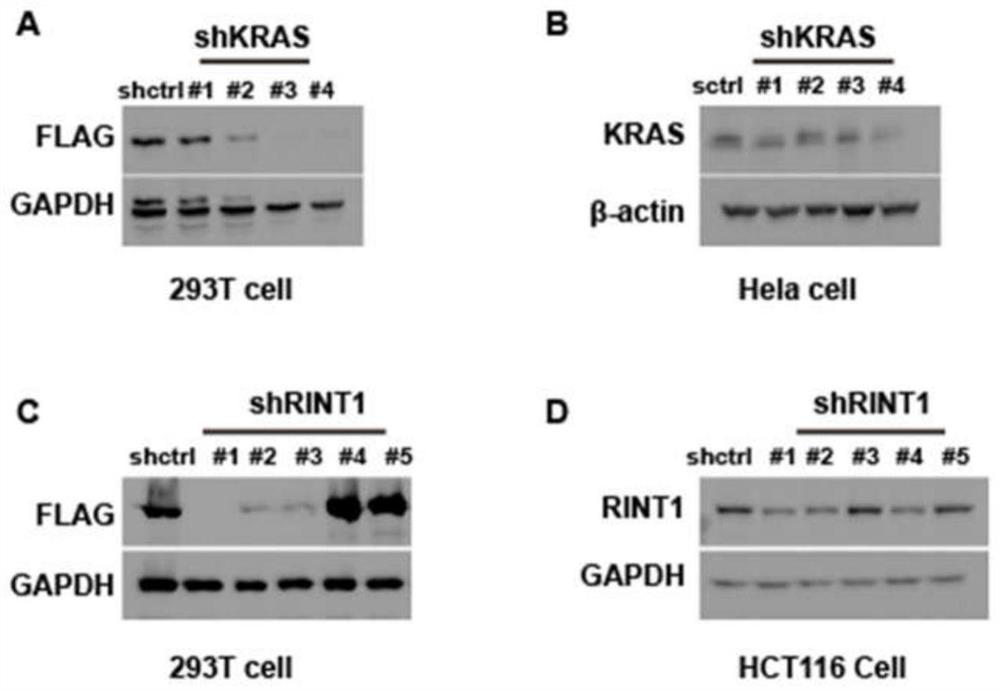
Method for rapidly identifying shRNA effectiveness
An effective and fast technology, applied in the field of molecular biology, can solve problems such as high cost, long time, and high risk of biological safety, and achieve the effect of reducing workload, maintaining consistency, and reducing labor costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0021] 1. Design and synthesize multiple shRNA sequences as candidate functional shRNAs according to the target gene sequence and the shRNA design rules of the PLKO.1 vector;
[0022] Specific implementation steps: log in to the shRNA design website, enter the name of the target gene, and search for candidate siRNA sequence fragments of the target gene. After the sequence specificity was verified by the blast comparison tool in the NCBI database, according to the design rules of PLKO.1shRNA (Forward oligo: 5'CCGG—21bpsense—CTCGAG—21bpantisense—TTTTTG3', Reverse oligo:5'AATTCAAAAA—21bp sense—CTCGAG—21bp antisense3') to complete the design of shRNA sequence fragments and send them to chemical synthesis.
[0023] 2. According to the vector construction steps, first dissolve the primers and configure them into an annealing reaction system, heat them in a constant temperature water bath at 95°C, and then cool and anneal naturally to form shRNA fragment dimers. At the same time, Ec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com