Method for detecting gene editing efficiency and gene editing mode and application of method
A gene editing and efficient technology, applied in biochemical equipment and methods, microbial measurement/inspection, instruments, etc., can solve the problems of large amount of high-throughput sequencing data, high equipment requirements, high false positive rate, etc., and achieve optimal statistics Analyzing the flow, resolving false positives, and streamlining the effect of the build process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
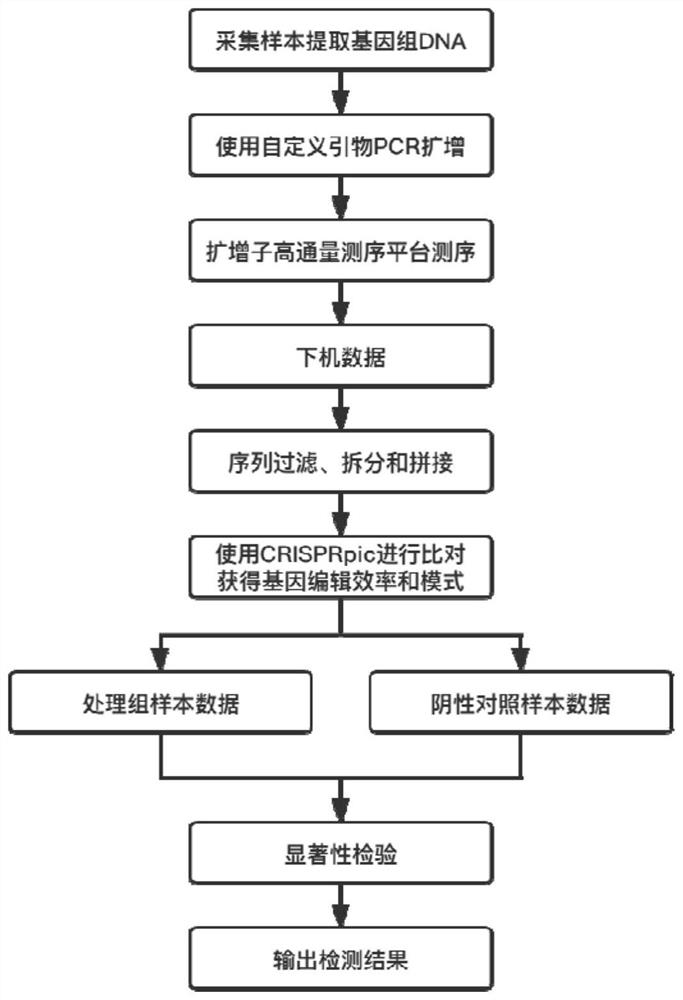
[0039] The method for detecting gene editing efficiency and gene editing mode comprises the following steps:
[0040] S1, extract the genomic DNA of the sample, design and amplify primers for the region to be tested;
[0041] S2, building a library for high-throughput sequencing, and filtering, splitting and splicing the data to obtain the sequence of the region to be tested;
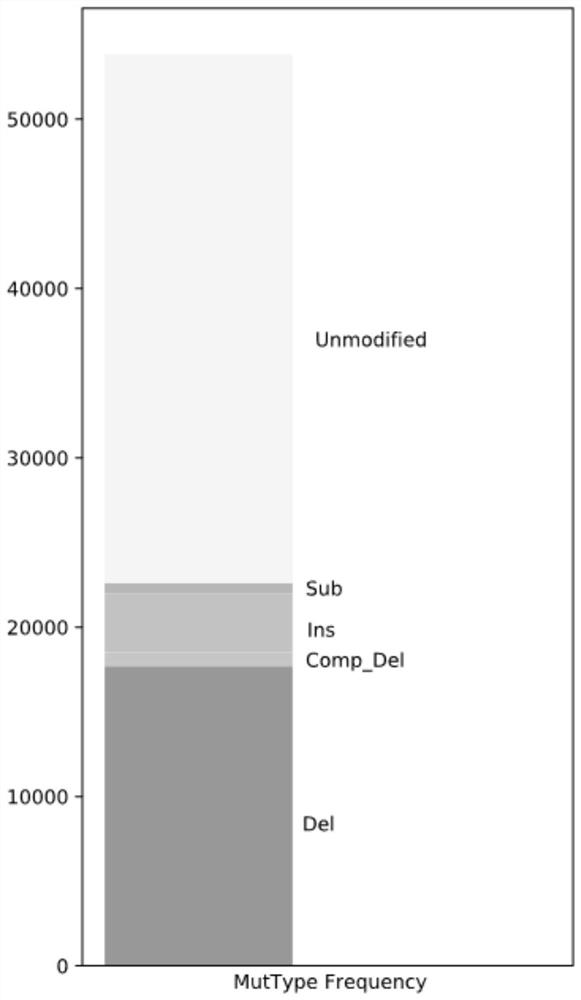
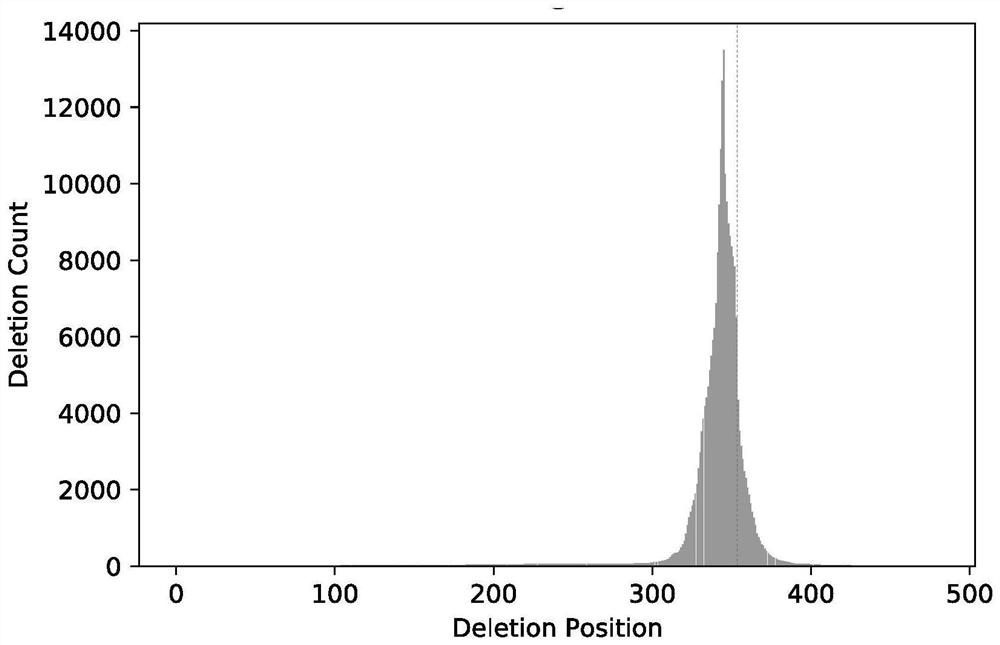
[0042] S3, comparing the sequence of the region to be tested with the genome reference sequence to obtain the gene editing efficiency and gene editing mode of each sample;
[0043] S4, performing a significance analysis on the gene editing efficiency of the samples in the treatment group and the samples in the control group, and outputting the corrected results.
[0044] Wherein, in the process of designing primers for the region to be tested in the step S1, the designed primers are amplification primers added with different adapter sequences, which can distinguish each sample when multiple samples are...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com