MSH6 gene with mutation at 12759 site and application thereofA technology of mutating genes and pathogenic genes, applied in the field of new hypophosphatemic rickets pathogenic genes and its detection kits, can solve the problems of non-absorption and metabolism research reports
Active Publication Date: 2021-03-19黄志玲View PDF5 Cites 0 Cited by - Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
The protein encoded by the MSH6 gene belongs to the mismatch repair protein, also known as G / T mismatch binding protein (G / T mismatch binding protein, GTBp), which has the function of participating in DNA base mismatch repair, and can participate in the recognition and binding to the mismatch site After the repair point, the repair process of the mismatch site is completed under the action of other mismatch repair-related enzymes. Studies have shown that this gene may play a role in the occurrence and development of tumors, but there is no research report showing that the gene It is associated with HR, and there is no research report on the relationship between this gene and the absorption and metabolism of phosphorus in the body Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine View moreImage
Smart Image Click on the blue labels to locate them in the text.Viewing ExamplesSmart Image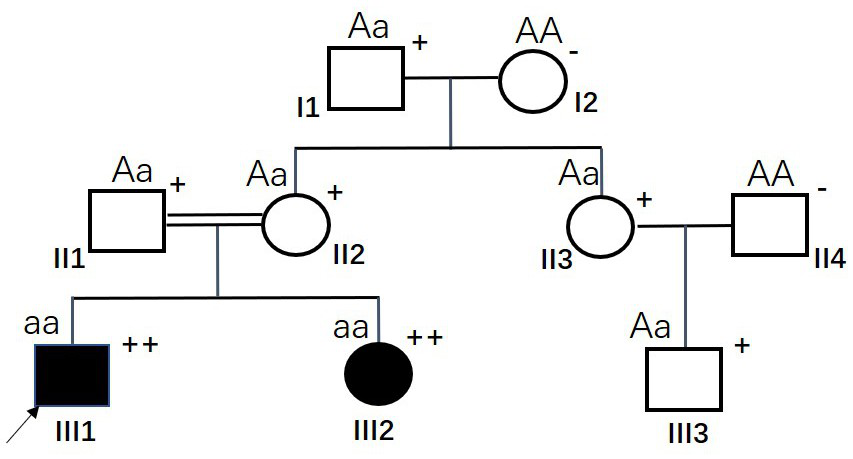
Examples
Experimental program Comparison scheme Effect test Embodiment 1
[0020] Example 1 found that the specific MSH6 mutation gene is the pathogenic gene of HR
[0021] figure 1This is the HR family with the earliest MSH6 gene mutation detected (family #1), II1 and II2 are consanguineous marriages (II1 is the only child and the parents have died). Among them, both siblings III1 and III2 showed symptoms of short stature and rickets, as well as hypophosphatemia and increased urinary phosphorus, and were clinically diagnosed as HR. Except for HR, III1 (the proband, marked by the arrow) was diagnosed with stage III colorectal cancer at the age of 24 in 2018. The tumor was surgically removed, but recurrence occurred six months after the operation. Family history investigation revealed that both III1 and III2 in #1 family were HR patients, and III1 was diagnosed with colorectal cancer, which has certain pathogenic genetic indications and tendencies. In order to guide the follow-up treatment of colorectal cancer patient III1, analyze the potential gen...
Embodiment 3
[0044] Embodiment 3 HR population sample verification
[0045] On the basis of Examples 1 and 2, the MSH6 gene mutation was further detected in the HR population. A total of 27 clinically confirmed HR patients (number S1-S27) were collected, including 16 male patients and 11 female patients, aged 26.3 years (4-57 years old). According to the PCR capture sequencing method in Example 1 (Table 1 and Table 2), under the condition of full informed consent of these HR patients, blood samples (peripheral venous anticoagulant blood 2ml) were collected for MSH6 gene mutation detection, the results are shown in Table 5 shown. Among them, only S1-S17 HR patients were detected to contain specific mutations of the MSH6 gene, while none of the S18-S27 HR patients were detected to contain any MSH6 gene mutations, indicating that there are other known or unknown gene mutations that can lead to HR. Occurs (such as the PHEX mutant gene on the X chromosome). In S1-S17 HR patients, only the g....
Embodiment 4
[0052] Application of embodiment 4 specific MSH6 mutant gene
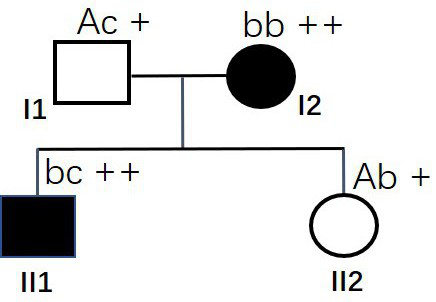
[0053] like Figure 8 The family shown (family #7), both husband and wife are normal persons, has given birth to a child with HR, and now plans to have another child and undergo genetic counseling. Blood samples from the husband and wife and the child with HR were collected for whole exome capture sequencing, and it was found that the father I1 had the g. [12759inTTAAG] mutation (heterozygous, genotype is Ac), while HR child II1 has MSH6 gene g.[12759inTTAAG] mutation and g.[12907inCAGC] mutation (compound heterozygous mutation, genotype is cd). These two MSH6 gene mutations were inherited from their mother and father respectively; except for the two specific mutations of the MSH6 gene, no other obvious gene mutations were detected in the family members. The phenotype was determined by the genotype in this family, and the phenotype was co-segregated with the genotype. Combined with the results of Examples 1-3, it...
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses an MSH6 mutant gene with g.[12759inTTAAG] mutation as a new pathogenic gene of HR. The mutation fragment sequence of the MSH6 mutant gene is shown as SEQ ID NO: 3. The diploidhomozygous genotype of the MSH6 mutant gene with the sequence shown as SEQ ID NO: 3 can cause human hypophosphorous rickets, and the diploid heterozygous genotype composed of any MSH6 mutant gene in the sequences shown as SEQ ID NO: 3, SEQ ID NO: 1, SEQ ID NO: 2 and SEQ ID NO: 4 can also cause human hypophosphorous rickets, and are autosomal recessive inheritance. On the basis, the invention provides two mutation detection kits based on PCR capture sequencing and conventional PCR and Sanger sequencing, which are of great significance for HR screening, diagnosis and fertility guidance, and areespecially conducive to completely eradicating the birth of HR children from the source. In addition, the disclosure of specific mutation of the MSH6 gene is also conducive to the exploration of HR pathogenesis and the development of therapeutic drugs and methods.Description
technical field [0001] The invention relates to the field of medical molecular biology, in particular to a novel hypophosphatemic rickets pathogenic gene and a detection kit thereof. Background technique [0002] Hypophosphatemic rickets (Hypophosphatemic Rickets, HR) is the main type of vitamin D-resistant rickets. Due to the weakened recovery of phosphorus in the renal tubules of patients with HR, hypophosphatemia and vitamin D metabolism disorders will occur, and at least lead to hypophosphatemia. Saltemia can develop into rickets or osteomalacia in severe cases, and it will also affect hearing and teeth. Children's HR mainly manifests as growth retardation, short stature, rickets, bone and tooth deformities, osteoporosis, walking weakness, etc.; adult HR mainly causes osteomalacia and bone and joint deformities. Biochemical examination showed that the blood phosphorus of HR patients was significantly lower, and the alkaline phosphatase was significantly higher, and the ...Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine Login to View More Application Information
Patent Timeline  Login to View More IPC IPC(8): C12N15/12C12Q1/6858C12Q1/6883CPCC12Q1/6858C12Q1/6883C12Q2600/156C12Q2531/113C12Q2535/122C12Q2535/101 Inventor 黄志玲 Owner 黄志玲
Login to View More IPC IPC(8): C12N15/12C12Q1/6858C12Q1/6883CPCC12Q1/6858C12Q1/6883C12Q2600/156C12Q2531/113C12Q2535/122C12Q2535/101 Inventor 黄志玲 Owner 黄志玲Features- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media Patsnap Eureka Blog Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com
A technology of mutating genes and pathogenic genes, applied in the field of new hypophosphatemic rickets pathogenic genes and its detection kits, can solve the problems of non-absorption and metabolism research reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Example 1 found that the specific MSH6 mutation gene is the pathogenic gene of HR
[0021] figure 1This is the HR family with the earliest MSH6 gene mutation detected (family #1), II1 and II2 are consanguineous marriages (II1 is the only child and the parents have died). Among them, both siblings III1 and III2 showed symptoms of short stature and rickets, as well as hypophosphatemia and increased urinary phosphorus, and were clinically diagnosed as HR. Except for HR, III1 (the proband, marked by the arrow) was diagnosed with stage III colorectal cancer at the age of 24 in 2018. The tumor was surgically removed, but recurrence occurred six months after the operation. Family history investigation revealed that both III1 and III2 in #1 family were HR patients, and III1 was diagnosed with colorectal cancer, which has certain pathogenic genetic indications and tendencies. In order to guide the follow-up treatment of colorectal cancer patient III1, analyze the potential gen...
Embodiment 3
[0044] Embodiment 3 HR population sample verification
[0045] On the basis of Examples 1 and 2, the MSH6 gene mutation was further detected in the HR population. A total of 27 clinically confirmed HR patients (number S1-S27) were collected, including 16 male patients and 11 female patients, aged 26.3 years (4-57 years old). According to the PCR capture sequencing method in Example 1 (Table 1 and Table 2), under the condition of full informed consent of these HR patients, blood samples (peripheral venous anticoagulant blood 2ml) were collected for MSH6 gene mutation detection, the results are shown in Table 5 shown. Among them, only S1-S17 HR patients were detected to contain specific mutations of the MSH6 gene, while none of the S18-S27 HR patients were detected to contain any MSH6 gene mutations, indicating that there are other known or unknown gene mutations that can lead to HR. Occurs (such as the PHEX mutant gene on the X chromosome). In S1-S17 HR patients, only the g....
Embodiment 4
[0052] Application of embodiment 4 specific MSH6 mutant gene
[0053] like Figure 8 The family shown (family #7), both husband and wife are normal persons, has given birth to a child with HR, and now plans to have another child and undergo genetic counseling. Blood samples from the husband and wife and the child with HR were collected for whole exome capture sequencing, and it was found that the father I1 had the g. [12759inTTAAG] mutation (heterozygous, genotype is Ac), while HR child II1 has MSH6 gene g.[12759inTTAAG] mutation and g.[12907inCAGC] mutation (compound heterozygous mutation, genotype is cd). These two MSH6 gene mutations were inherited from their mother and father respectively; except for the two specific mutations of the MSH6 gene, no other obvious gene mutations were detected in the family members. The phenotype was determined by the genotype in this family, and the phenotype was co-segregated with the genotype. Combined with the results of Examples 1-3, it...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com
