Optimization method for output result of Kraken2 software and method for identifying species types in sample
A technology for outputting results and optimizing methods, applied in the biological field, can solve the problem of high false positive rate, improve the accuracy of results and avoid false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Analysis of simulated sequencing data for 40 species
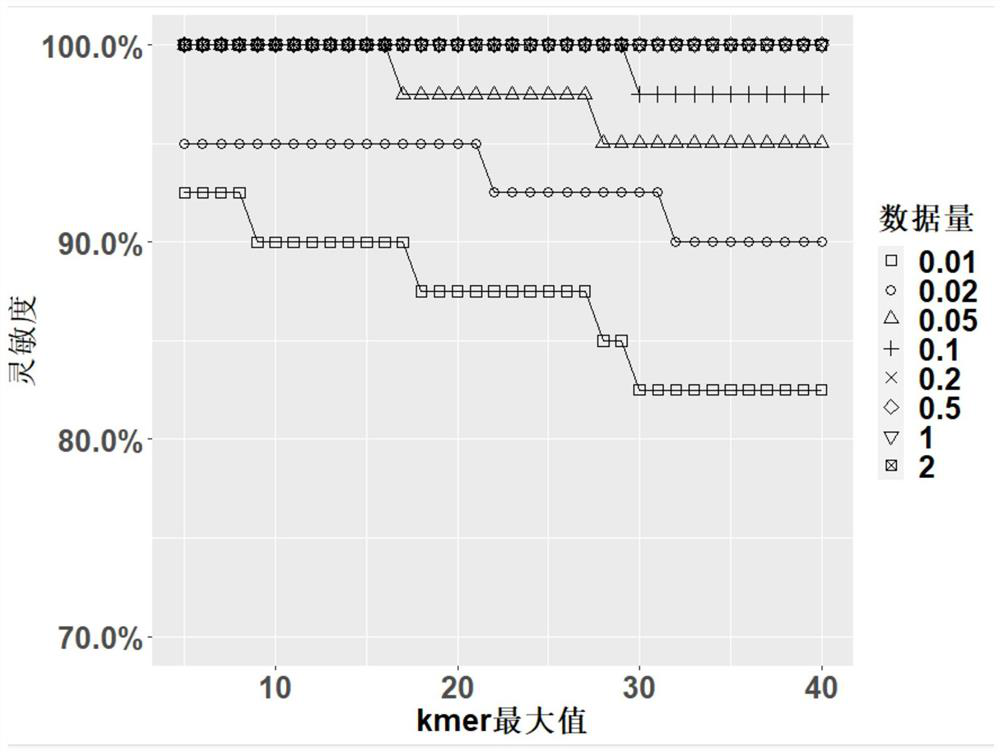
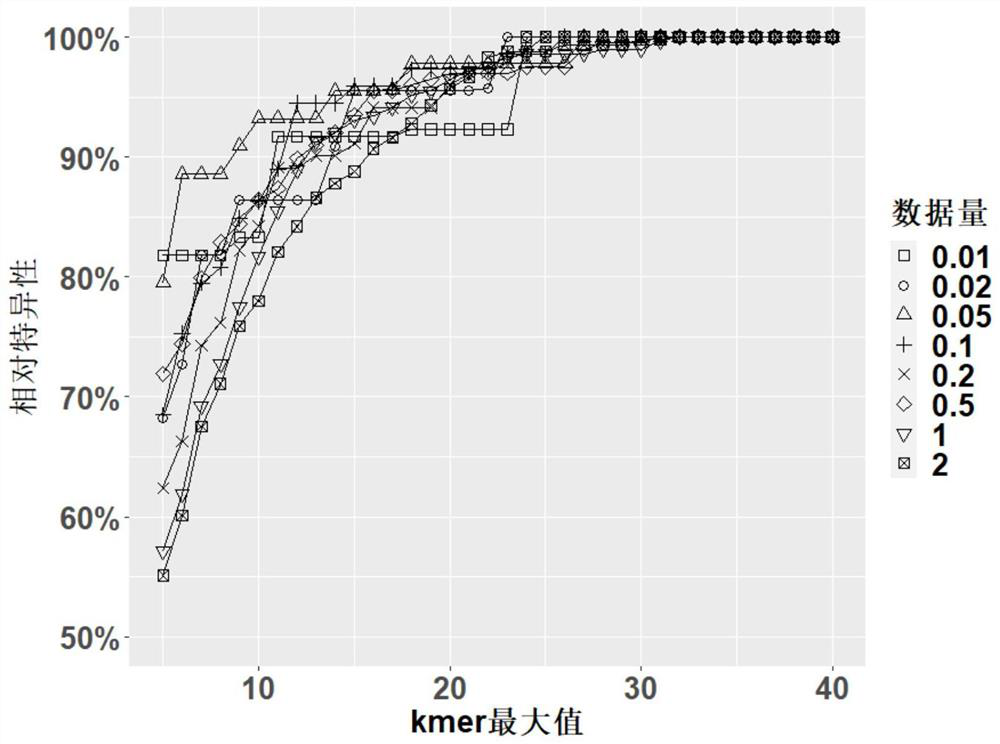
[0038] First, the genomes of 40 representative species (Table 1) were randomly selected from the database, covering eukaryotes, bacteria, and viruses, including some species with close relatives. Then use the sequencing data simulation tool ART to generate double-ended 75bp sequencing data and run Kraken2 to identify these species. Finally, the results of Kraken2 are screened with the two parameters kmermax and kmersum, and the sensitivity, relative specificity and accuracy under different data volumes are calculated.
[0039] Sensitivity is the ratio of the number of detected true positive species X to the number of theoretical true positive species (40), ie X / 40.
[0040] Since there is no true negative value in the original data, in order to evaluate the results, we use the original results of Kraken2 as the basis. The false positive species in this result are regarded as the theoretical true negative...
Embodiment 2
[0062] Example 2: ZymoBIOMICS TM Analysis of sequencing data of MICROBIAL Community Standard (Catalog No.D6300)
[0063] Will ZymoBIOMICS TM The MICROBIAL Community Standard standard (Table 5) was added to triple-distilled water for library construction and sequencing. After quality control of the paired-end 75bp sequencing data and removal of human sources, Kraken2 was run for species identification and the parameters kmermax and kmersum were used. filter. Finally, the sensitivity, relative specificity and correct rate under different data volumes are calculated.
[0064] Table 5 ZymoBIOMICS TM MICROBIAL Community Standard
[0065]
[0066] Sensitivity is the ratio of the number of detected true positive species X to the number of theoretical true positive species (10), ie X / 10.
[0067] Since there is no true negative value in the original data, in order to evaluate the results, we use the original results of Kraken2 as the basis. The false positive species in thi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com