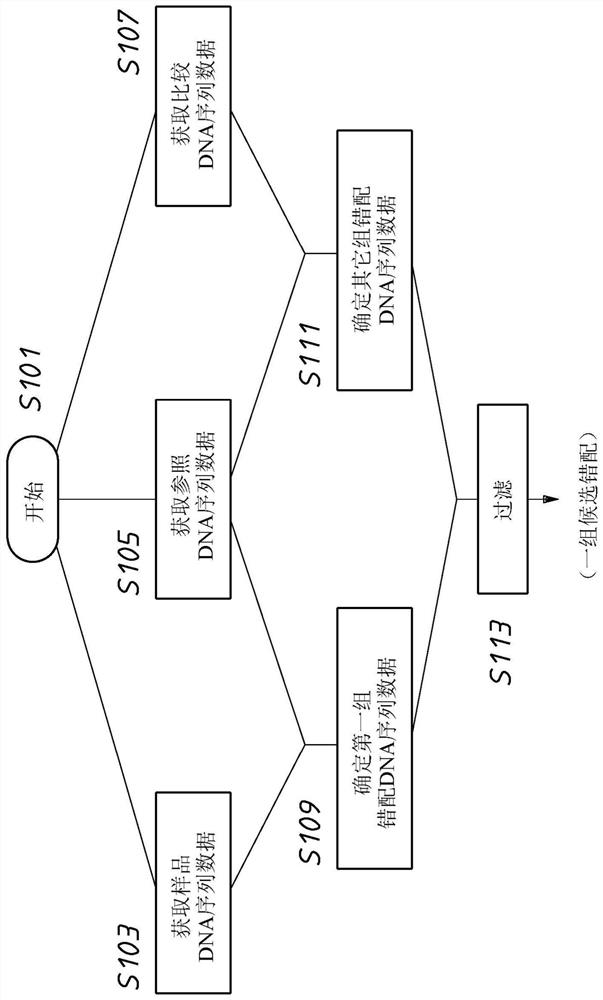
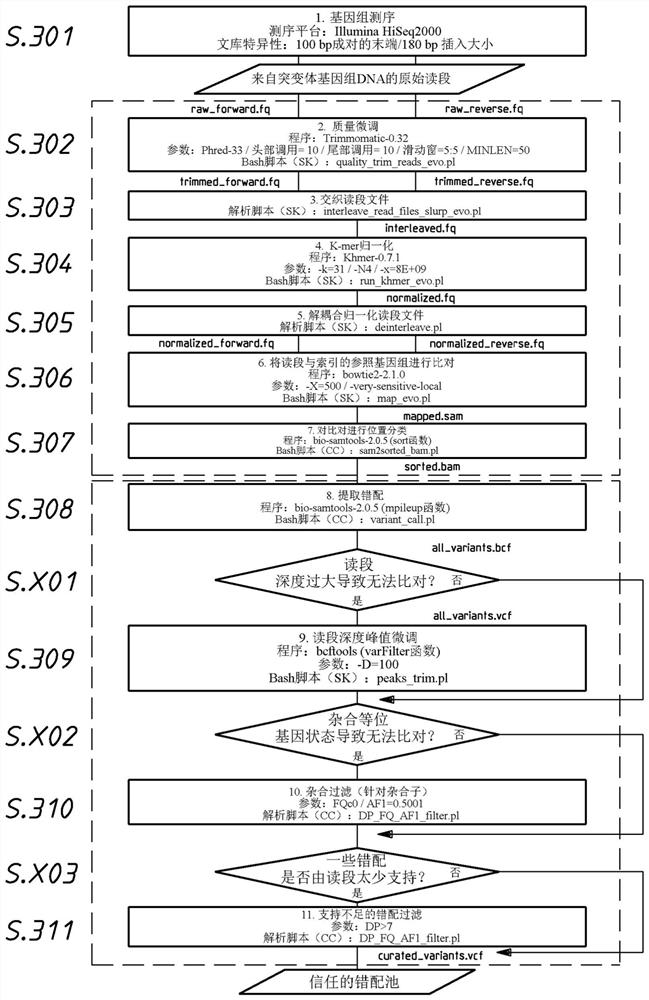
A method or system for identification of a causative mutation causing a phenotype of interest in a test sample
A test sample, purpose technology, applied in the field of identification of disease-causing mutations or systems that lead to the phenotype of interest in the test sample, can solve the problems of costing money, requiring time, limiting pipeline throughput, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0530] Example 1: Mutations found in the gene for the plant-enhancing protein RHO GTPASES that impair fertility (Case B)
[0531] Several independent mutant lines were generated by irradiating N. polymorpha with UV-B. The mutant lines were divided into two phenotypic groups: some with straight rhizoids ( Figure 8 A) and intact epidermis ( Figure 9 A), some with wavy rhizoids ( Figure 8 B) and stretched epidermis ( Figure 9 B).
[0532] We aimed to identify causative mutations in UV4.32 mutant lines with wavy rhizoids and elongated cuticles. DNA was extracted from UV4.32 mutants with wavy rhizoids and elongated cuticles using whole plants as samples and standard DNA phenol-chloroform-IAA. The genome of UV4.32 and seven independent mutant lines with straight rhizoids and intact epidermis were sequenced using Illumina's HiSeq-2000 platform technology.
[0533]Raw reads were fine-tuned for quality using Trimmomatic-0.32 and normalized using Khmer0.7.1 with a k-mer size o...
example 2
[0542] Example 2: A mutation leading to resistance to chlorsulfuron was found in the acetolactate synthase gene (case A)
[0543] Polymorpha polymorpha was irradiated with UV-B and seven independent mutant lines resistant to the herbicide chlorsulfuron-methyl were identified. Chlorsulfuron resistance was determined from surviving Diana polymorpha plants after two weeks of exposure to a lethal dose of chlorsulfuron (0.1 ppm dose, a dose sufficient to kill 100% of wild-type plants).
[0544] Since all mutant plants had the same phenotype of chlorsulfuron resistance, we assumed that they each contained the same pathogenic mutation. The chlorsulfuron-resistant mutants were compared to a reference genome of over 100,000 mismatches identified individually, and we first filtered for mismatches that were also present in the M0 wild-type genome ( Figure 11 , the 2 leftmost scatter boxes).
[0545] To test the efficiency of the allelism-based version of the pipeline, we applied it to...
example 3
[0550] Example 3: A mutation leading to resistance to chlorsulfuron was found in the acetolactate synthase gene (AB case)
[0551] To increase the power of the pipelines exemplified in Examples 1 and 2, we combined two approaches: In this example of the pipeline, the Find the causative mutation in the mismatch group that exists.
[0552] Using 3 chlorsulfuron-sensitive mutagenized lines, we filtered 4 of 11 chlorsulfuron-resistant-specific mismatches previously identified as consistent with the expected mutation signature and in the coding sequence of the gene , which ended up leaving us with only 4 candidate mutations (Table 4) that were predicted to result in changes in the amino acid sequence of the protein.
[0553] This represents a 20% to 30% increase in pipeline capacity compared to the pipeline exemplified in Example 2 alone. Because the capacity of the pipelines in Examples 1 and 2 increases with the number of allelic and non-allelic minus lines, respectively, we pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com