Method for rapidly detecting E4P production capability of to-be-detected strain and biosensor used in method
An affinity, a14 technology, applied in biological testing, chemical instruments and methods, instruments, etc., can solve problems such as time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
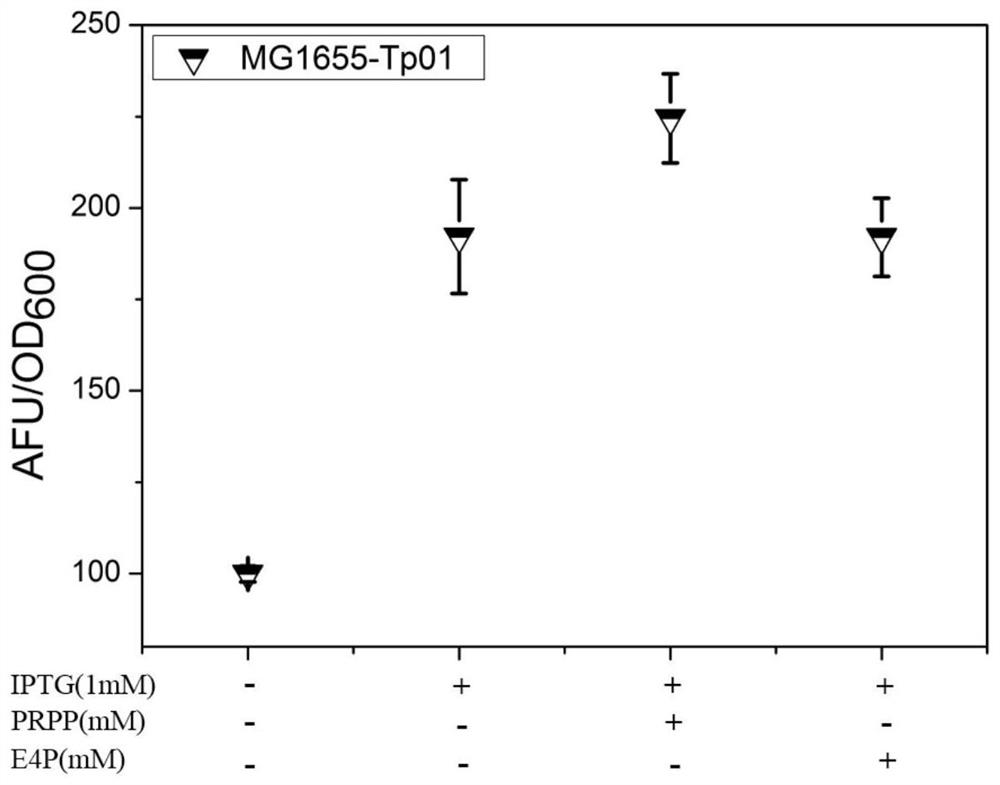
[0117] Embodiment 1, construction and testing of sensor plasmid Tp01
[0118] 1. Construction of sensor plasmid Tp01
[0119] Ligand-binding proteins for biosensors should have high affinity and specificity for ligands. HGPRT protein uses PRPP and purine as substrates, and catalyzes the transfer of phosphoribosyl sugar on PRPP to purine, so that purine phosphoribosylation forms purine nucleotides. The Km value of human HGPRT protein to PRPP is very low (<0.1mM). Through random mutation, Hemalatha et al. obtained a HGPRT mutant F36L, which further reduced the Km (<1 μM) of PRPP and could still maintain more than 80% activity at 60°C (Raman, J., Sumathy, K. , Anand, R.P. & Balaram, H.A non-active sitemutation in human hypoxanthine guanine phosphoribosyltransferase expands substrate specificity. Archives of biochemistry and biophysics 427, 116-122(2004).). Therefore, the inventors of the present invention introduced the F36L mutation point on the basis of the human HGPRT prote...
Embodiment 2
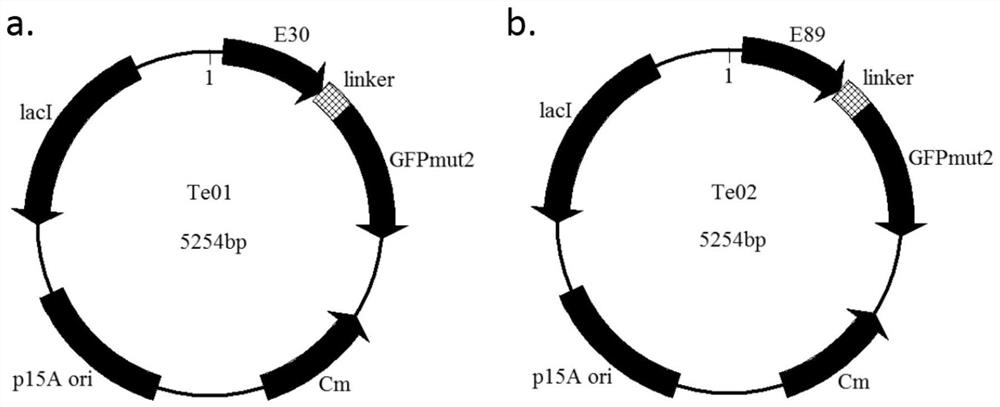
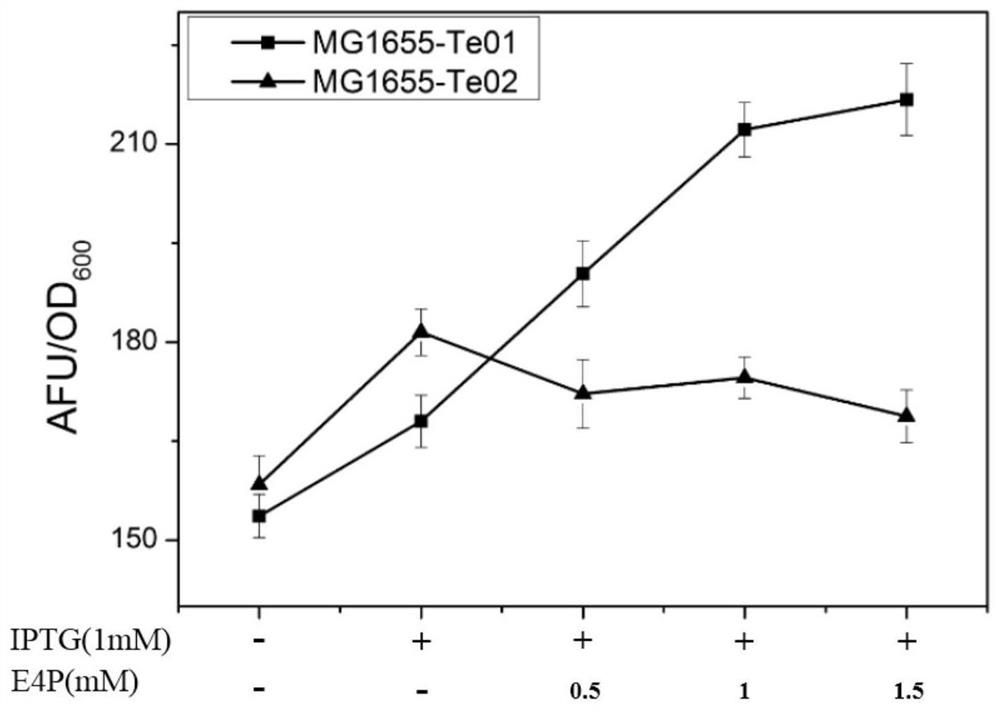
[0145] Embodiment 2, construction and testing of sensor plasmid Te01 and sensor plasmid Te02
[0146] 1. Obtaining protein E30 and protein E89
[0147] On the basis of protein T0, the inventors of the present invention docked and matched the ligand E4P to the binding pocket region, simulated mutations on the residues around the ligand by computational means, and evaluated the mutation effect to stabilize the ligand and increase the binding capacity. The main reference method is to obtain protein E30 and protein E89 through computational optimization.
[0148] The amino acid sequence of protein E30 is shown in SEQ ID NO:6. The protein E30 is codon-optimized and gene-synthesized to obtain the gene encoding the protein E30 (ie, the E30 gene).
[0149] The amino acid sequence of protein E89 is shown in SEQ ID NO:7. The protein E89 is codon-optimized and gene-synthesized to obtain the gene encoding the protein E89 (ie, the E89 gene).
[0150] 2. Construction of sensor plasmid T...
Embodiment 3
[0178] Embodiment 3, Rational transformation of E30 protein
[0179] Compared with T0 protein, protein E30 has a certain responsiveness to E4P molecule. Furthermore, in order to improve the sensitivity of the protein to respond to E4P, a series of rational designs were carried out on the E30 protein. The design principle is mainly based on the mechanism of sensor action: in the absence of ligand, the ligand-binding protein will be degraded by the intracellular degradation mechanism, so the fluorescent protein cannot exist stably; in the presence of the ligand, the ligand-binding The protein will bind to the ligand, the ligand-binding protein can exist stably in the cell, and the fluorescent protein can be detected. Therefore, the computer simulation analysis of the three-dimensional structure of protein E30 found that there are multiple loop regions in the E30 protein, which are closely related to the protein binding pocket. The modification of the loop region near the active...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com