Non-coding small RNA molecular spectrum related to tumor lactic acid microenvironment and application thereof
A technology of small molecules and lactic acid, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, etc., and can solve the problems of difficult acquisition of pathological specimens, infection, necrosis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0101] Example 1: Screening and Identification of Lactic Acid Microenvironment Responsive miRNA Molecules
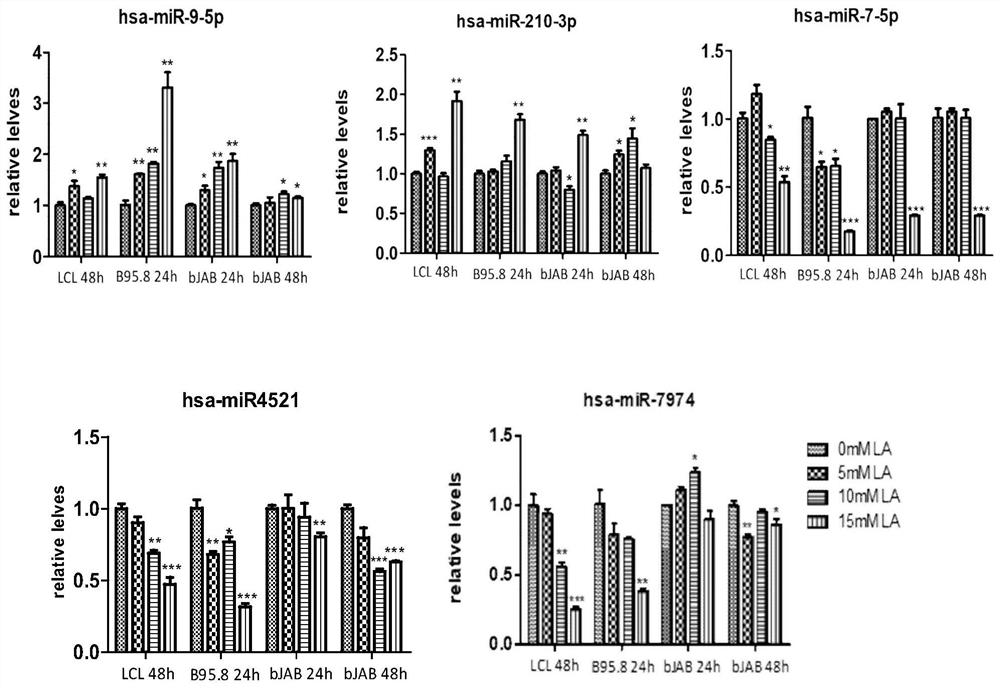
[0102] Using stem-loop qRT-PCR, in EBV-positive cell lines (LCL, B95.8) and EBV-negative cell lines (BJAB), the expression levels of miRNA molecules under different concentrations of lactic acid were detected, and a group of lactic acid-responsive and A miRNA expression profile with a stable trend. This group of miRNAs is hsa-miRNA-7974, hsa-miRNA-7-5p, hsa-miR-4521, hsa-miRNA-9-5p, hsa-miRNA210-3p ( image 3 ).
[0103] Detection of miRNA-stem-loop real-time quantitative PCR: a stem-loop reverse transcription primer is designed separately for each miRNA, and the 5' end of the stem-loop RT primer specifically binds to the 3' end of the miRNA molecule. Under the action of the enzyme, the lengthened miRNA first-strand cDNA is obtained. The obtained reverse transcription product is subjected to qPCR reaction under the action of qPCR specific upstream primer and downstrea...
Embodiment 2
[0136] Example 2: Lactic acid microenvironment responds to 5'-tiRNA HisGUG screening and identification of
[0137] A 5'-tRNA moiety: 5'-tRNA was screened out in EBV-positive cell lines (LCL1 and B95.8) by fluorescent quantitative PCR HisGUG , its expression has a certain trend of change with the increase of lactic acid concentration. At the same time, the expression level of ANG molecules upstream of the cleaved tRNA was detected, and the results showed that the expression level of ANG also changed with the increase of lactic acid concentration ( Figure 7 ). This suggests that 5'-tRNA HisGUG Lactic acid plays a certain role in promoting the occurrence of B-cell lymphoma, and has potential clinical value in the diagnosis and prognosis of EBV-related B-cell lymphoma.
[0138]Detection and quantitative analysis of tRNA-half molecules: The structural feature of 5-tiRNA is that there are 2', 3'-cyclic phosphate groups at the 3' end. In the first step, T4 PNK treats the total ...
Embodiment 3
[0156] Example 3: Detection of expression levels of hsa-miR-9-5p, hsa-miR-210-3p, hsa-miR-7-5p, hsa-miR4521, hsa-miR-7974 in 8 lymphoma samples
[0157] Eight clinical samples (PBMC) of lymphoma and other diseases with EBV negative and normal LDH (lactate dehydrogenase) levels ( Figure 7 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com