SARS-CoV-2 lymphocyte antigen epitope peptides and application thereof
A sars-cov-2 and epitope peptide technology, applied in the field of medical immunology and infectious diseases, can solve the problems of restricting the development of vaccines and therapeutic preparations, restricting the study of immune pathology and immune protection mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
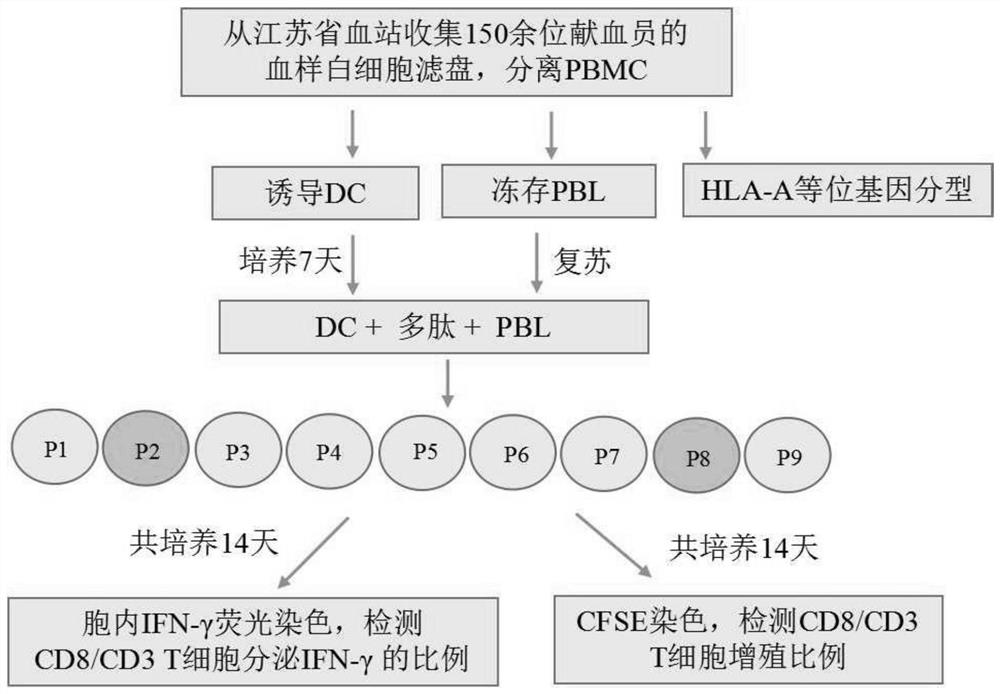
[0037] Embodiment 1: In some examples of the present invention, a method for predicting and identifying the amino acid sequence of a SARS-CoV-2 protein T cell epitope peptide is provided, and the specific steps are as follows:
[0038] 1. Virtual prediction of T cell epitope peptides restricted by HLA-A molecules from 5 SARS-CoV-2 proteins
[0039] Select 5 kinds of SARS-CoV-2 proteins, for example, E protein (envelope protein), M protein (membrane protein), N protein (nucleocapsid protein), S protein (spike protein) and RdRp protein (RNA-dependent RNA polymerase ). Then use SYFPEITHI, EPIJEN, ConvMHC, IEDB (including ANN, Consensus, NetMHCpan, SMM, SMMPMBEC) and other epitope peptide prediction tools to predict HLA-A0201, A1101, A2402, A3101, A0206, A0207, A3303, A3001, A0203, A1102, A0301, A0101, A2601 and other molecularly restricted T cell epitope peptides for each of the above proteins were virtually predicted.
[0040] The two ends of the antigen-binding groove of the ...
Embodiment 2
[0099] Example 2: In some examples of the present invention, part of the epitope peptides can be synthesized from the above 164 amino acid sequences, namely the T cell epitope peptide sequences shown in SEQ ID NO: 1-164, to prepare a mixed polypeptide vaccine. As an example, the preparation steps are as follows:
[0100] 1. Prepare the mixed peptide pool and adjuvant first
[0101] 1) Using the T cell epitope peptide sequences of SEQ ID NO: 1-164, synthesize 31 kinds of epitope peptides that can be presented by HLA-A2 molecules. Peptides can be synthesized by Suzhou Qiangyao Biotechnology Co., Ltd., with a purity of >95%, 4mg / peptide, 1mg per tube, and stored at -800°C for later use;
[0102] 2) Dissolving peptides: Take 1 tube (1 mg) of each peptide and store it at room temperature for pre-warming. Dissolve it with 20 μL DMSO, then add 180 μL PBS to make the peptide storage concentration 5 μg / μL, store at -80°C for later use.
[0103] 3) In this example, for example, the fo...
Embodiment 3
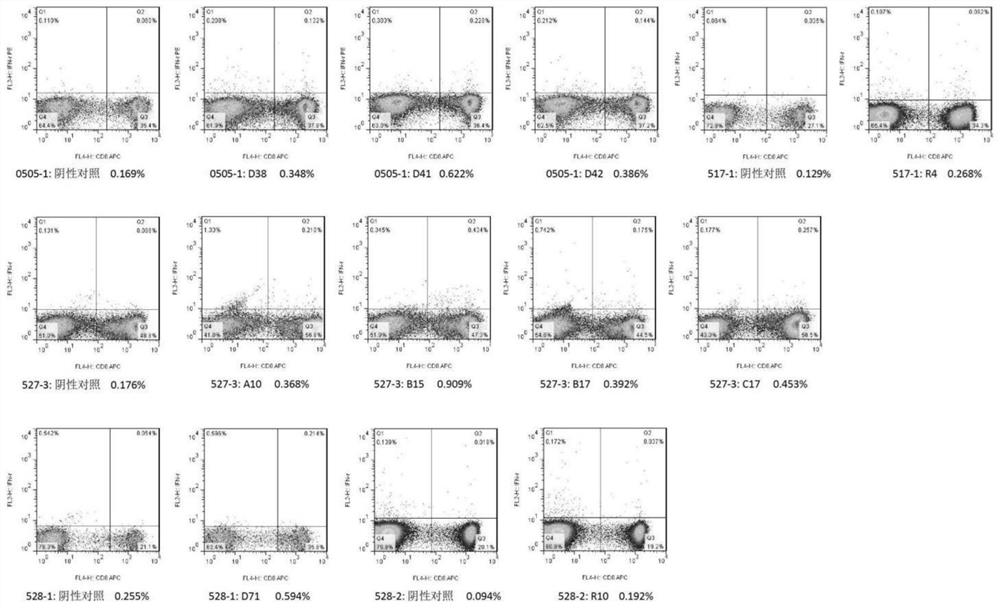
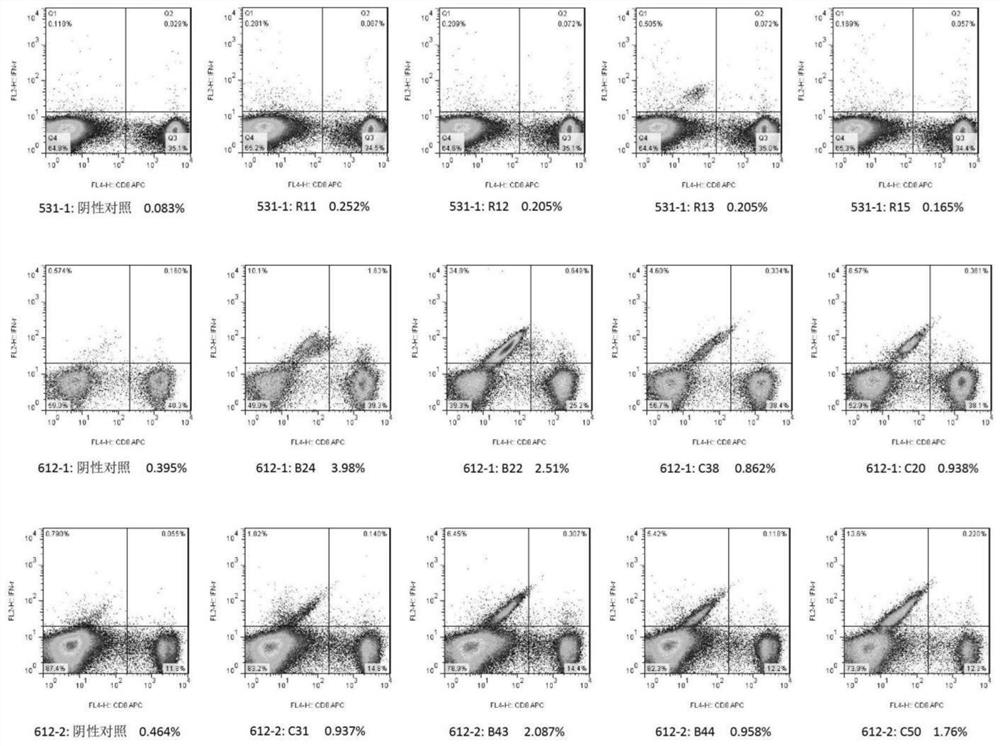
[0147] Example 3: In some examples of the present invention, epitope peptides were synthesized using the T cell epitope peptide sequences described in SEQ ID NO: 1-164, 10 peptide libraries were formed, and ELISPOT method was established to detect healthy blood donors The number of specific T cells that cross-react with the T cell epitope peptide of the SARS-CoV-2 protein in the population verifies the feasibility of using the above-mentioned expressed peptides to prepare reagents to detect the immune function of SARS-CoV-2 specific T cells. The specific steps can be as follows:
[0148] 1) Divide the above 164 epitope peptides into 10 peptide libraries according to the type of source protein, in which the epitope peptides of each protein are divided into 2 peptide libraries (23 epitope peptides for E protein, 36 epitope peptides for M protein , 25 kinds of epitope peptides of N protein, 41 kinds of epitope peptides of S protein, 39 kinds of epitope peptides of RdRp), each pep...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com