Primer composition for detecting SARS-CoV-2 and application thereof
A primer composition, sars-cov-2 technology, applied in the field of genetic engineering, can solve the problems of lack of SARS-CoV-2, insufficient scientific knowledge, accuracy and analysis specificity risks, and achieve rapid detection, good accuracy sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] This embodiment provides a SARS-CoV-2 detection kit and its use method, said kit includes the primer composition shown in SEQ ID No.1~12, the sequencing shown in SEQ ID No.7,9,11 Primers, reagents for PCR reaction and Sanger sequencing reagents, among which the reagents for PCR reaction include Baorui Bio’s reverse transcription amplification Mix, Baorui Bio’s mixed enzyme (including hot start Taq enzyme, reverse transcriptase, UNG enzyme), QIAGEN 10×buffer of QIAGEN, 5Q solution of QIAGEN, 25mM Mg of QIAGEN 2+ , 25 mM dNTPs from QIAGEN and H 2 O, the reagent for Sanger sequencing is Thermo Fisher's BigDye Terminatorv3.1 Cycle Sequencing Kit (including: BigDye, 5×seq Buffer).
[0096] The methods of use include:
[0097] (1) Collect nasopharyngeal swab samples;
[0098] (2) Nucleic acid was extracted by magnetic bead method;
[0099] (3) Use the primer combination whose nucleotide sequence is shown in SEQ ID No.1~12 to carry out the amplification reaction,
[0100]...
Embodiment 2
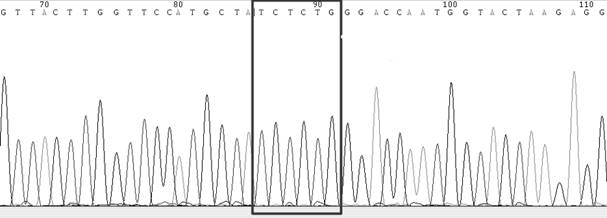
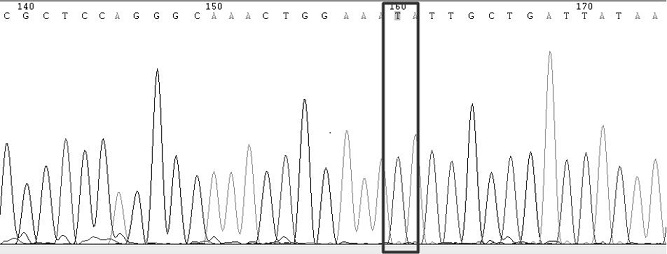
[0123] In this example, the kit and method described in Example 1 are used to detect SARS-CoV-2 (source of the sample: the remaining nucleic acid samples from human specimens detected by Kaipu Medical Laboratory), specifically for SARS-CoV-2 S protein Gene mutations g.21765_21770 del (p.HV 69_70del), g.A22812C (p.K417T), g.G22813C / T (p.K417N), g.G23012A (p.E484K), g.A23063T (p.N501Y) , g.A23403G (p.D614G) and g.C23604A (p.P681H) were detected.
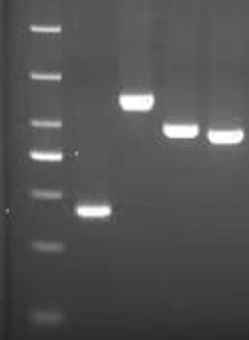
[0124] As shown in Table 5, SEQ ID NO:1,2 and SEQ ID NO:7,8 are respectively the primers of the first round of amplification and the second round of amplification of the g.21765_21770 del (p.HV69_70del) segment, first Carry out the first round of amplification with the primer shown in SEQ ID NO:1,2, and carry out the second round of amplification with the primer shown in SEQ ID NO:7,8, obtain No. 1 amplification product, likewise, with SEQID NO The first round of amplification is carried out with primers shown in 3 and 4, and the seco...
Embodiment 3
[0126] This example uses the kit and method described in Example 1 to detect SARS-CoV-2 (sample source: RNA sample of pseudovirus, pseudovirus purchased from Guangzhou Aiji Biotechnology Co., Ltd.), specifically for SARS-CoV -2 S protein gene mutations g.21765_21770 del (p.HV 69_70del), g.A22812C (p.K417T), g.G22813C / T (p.K417N), g.G23012A (p.E484K), g.A23063T ( p.N501Y), g.A23403G (p.D614G) and g.C23604A (p.P681H) were detected.
[0127] As shown in Table 5, SEQ ID NO:1,2 and SEQ ID NO:7,8 are respectively the primers of the first round of amplification and the second round of amplification of the g.21765_21770 del (p.HV69_70del) segment, first Carry out the first round of amplification with the primer shown in SEQ ID NO:1,2, and carry out the second round of amplification with the primer shown in SEQ ID NO:7,8, obtain No. 1 amplification product, likewise, with SEQID NO The first round of amplification is carried out with primers shown in 3 and 4, and the second round of am...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com