Construction method and application of fucose transferase 8 (FUT8) function-deleted cell strain
A technology of fucosyl transferase and function loss, which is applied in the field of molecular biology and can solve problems such as reducing the production and quality of antibody drugs and growth pressure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] The selection of embodiment 1 mutation site
[0075] In the present invention, the inventors found that H363, R365, D368, K369, E373, Y382, D409, D453 and S469 are all active sites of FUT8, but not all mutations at all sites are operable.
[0076] The requirements for screening suitable active sites in the present invention are based on the following conditions:
[0077] 1. Since CRISPR technology relies on the positioning of PAM (protospacer adjacent motif), this requires that the designed gRNA (guide RNA) must target the sequence near PAM;
[0078] 2. The base editing tool can only effectively realize the conversion of specific bases in the active window of the gRNA-targeted sequence at a fixed distance from the PAM sequence;
[0079] 3. Due to the degeneracy of codons, nonsense mutations may occur.
[0080] Based on the screening of the above three conditions, the inventors found three operable sites R365, D368 and D453.
[0081] The FUT8 protein sequence is as fo...
Embodiment 2
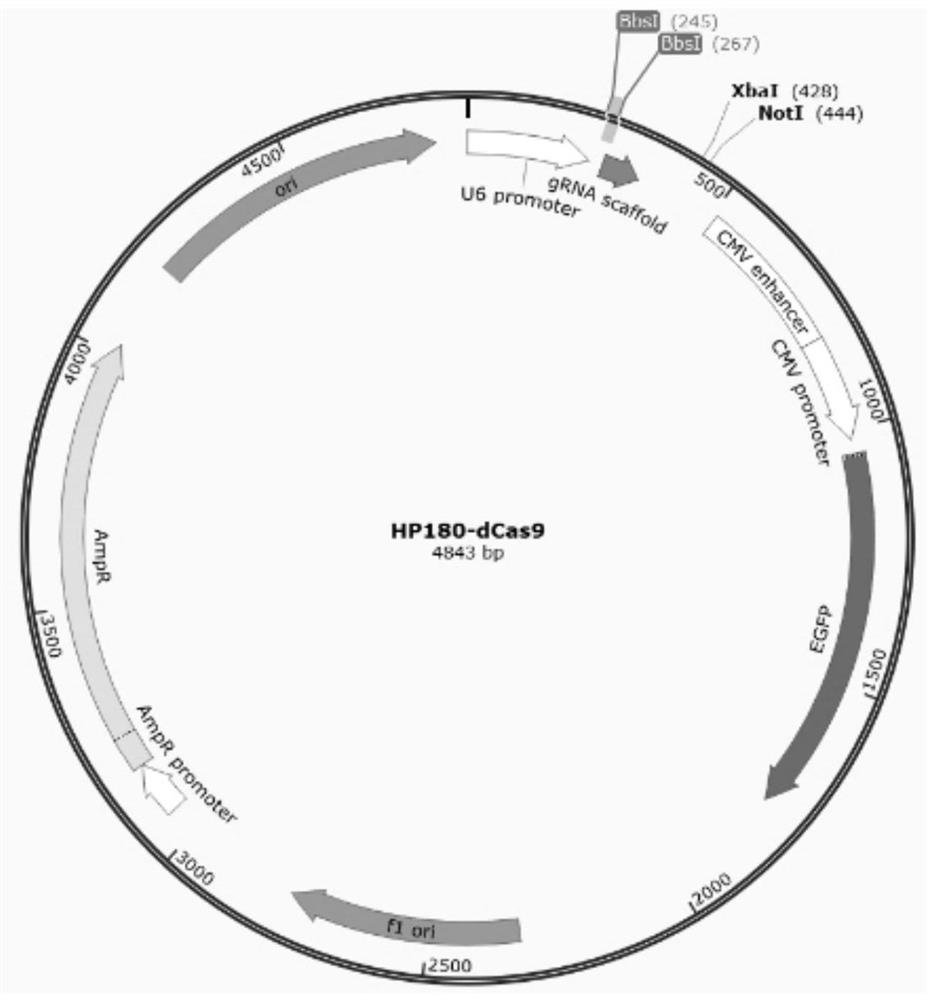
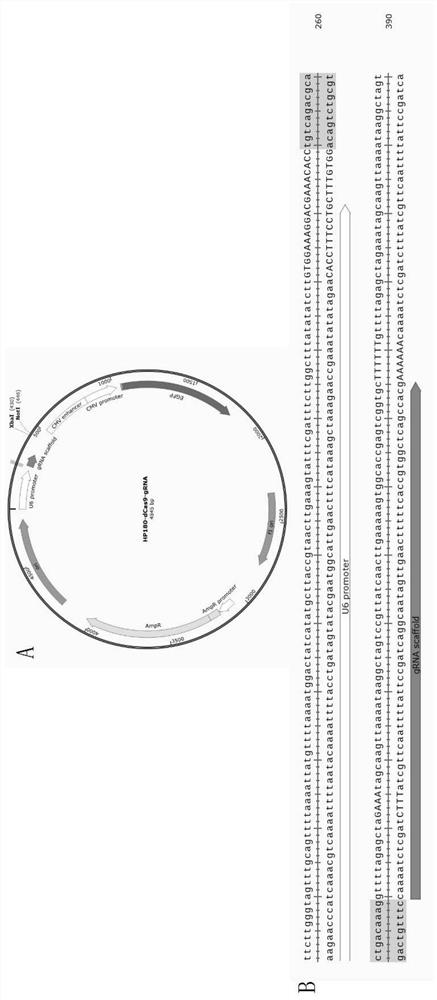
[0096] Embodiment 2 constructs plasmid
[0097] 1. Synthesize a targeting sequence DNA fragment with a restriction endonuclease BbsI site as follows:
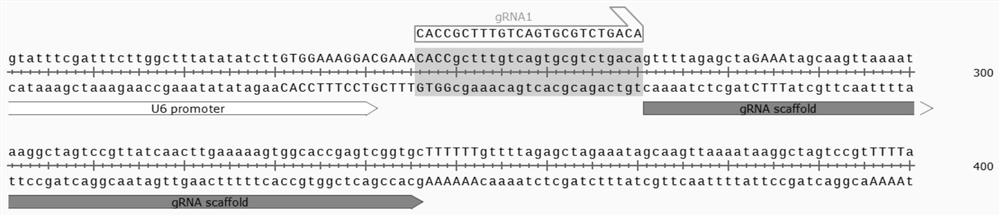
[0098] For gRNA1:
[0099] Forward: 5'-CACCGCTTTGTCAGTGCGTCTGACA-3' (SEQ ID NO.11)
[0100] Reverse: 5'-AAACTGTCAGACGCACTGACAAAGC-3' (SEQ ID NO.12)
[0101] For gRNA2:
[0102] Forward: 5'-CACCGTCAGACGCACTGACAAAGT-3' (SEQ ID NO.13)
[0103] Reverse: 5'-AAACACTTTGTCAGTGCGTCTGAC-3' (SEQ ID NO.14)
[0104] For gRNA3:
[0105] Forward: 5'-CACCTGTCAGACGCACTGACAAAG-3' (SEQ ID NO.15)
[0106] Reverse: 5'-AAACCTTTGTCAGTGCGTCTGACA-3' (SEQ ID NO.16)
[0107] For gRNA4:
[0108] Forward: 5'-CACCGGATATACACTTTTCTCTCCC-3' (SEQ ID NO.17)
[0109] Reverse: 5'-AAACGGGAGAGAAAGTGTATATCC-3' (SEQ ID NO.18)
[0110] The oligonucleotide chains of the above sequences were synthesized, and the forward and reverse strands of each group were mixed to make the final concentration 50 μM. Denatured at 94°C for 5 minutes, annealed at 50°C to obtai...
Embodiment 3
[0130] Example 3 transfection of CHO engineered cells
[0131] Co-transfect the HP180-dCas9 plasmid encoding gRNA (HP180-dCas9-gRNA1, 2, 3, 4) and the base editor plasmid (ABEmax or BE3) into CHO engineered cells, the process is as follows:
[0132] 1. Preparing Cells
[0133] at 37°C, 5% CO 2 CHO cells were cultured with CHO Fusion medium (containing 2% Glutamax and 1% penicillin and streptomycin (P / S) double antibody) in a cell culture box with a certain concentration, and after they grew to the logarithmic growth phase, count the cells Density, take the number of cells as 2×10 6 The cell solution was placed in a 15ml centrifuge tube, centrifuged at 100g for 5min, the supernatant was discarded, and then the cells were resuspended with 1.8ml Opti MEM medium (containing 2% GlutaMax and 1% P / S double antibody).
[0134] 2. Incubate the Transfection Reagent
[0135] After vortexing the FectoPRO reagent (purchased from Polyplus Transfection Company) for 5 seconds, take 2 μL i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com