Drop-off ddPCR method and kit for quantitatively detecting U2AF1 gene mutation
A quantitative detection and kit technology, applied in the biological field, can solve the problems of low sensitivity, real-time fluorescence quantitative PCR cannot perform absolute quantification, single mutation type, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
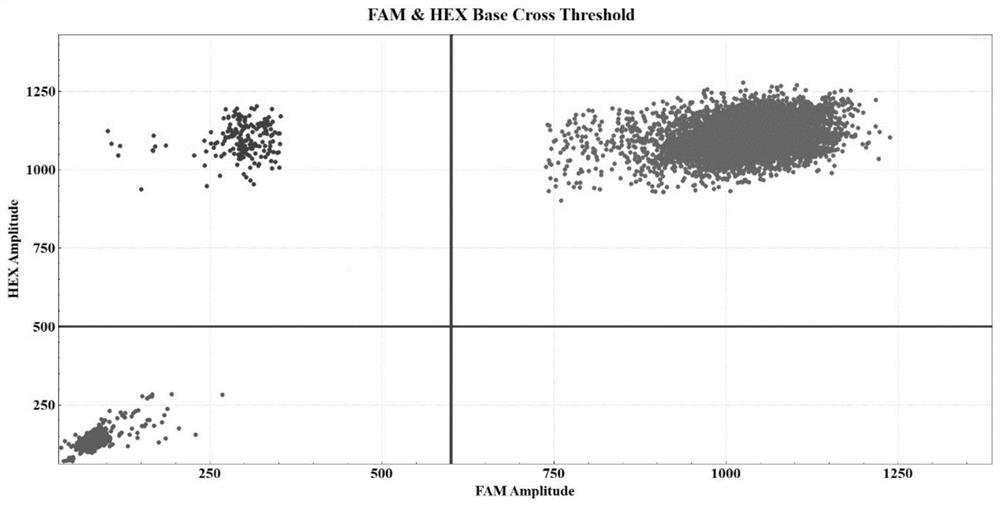
[0054] Taking the bone marrow samples of patients with U2AF1 gene Q157P mutation as an example, the detection method is described in detail.
[0055] 1. Isolation of bone marrow mononuclear cells and extraction of DNA
[0056] (1) Add 5ml of erythrocyte lysate to the bone marrow sample, mix well, and let stand for 1min;
[0057] (2) Centrifuge at 12000×rpm for 20sec, discard the supernatant and keep the precipitate;
[0058] (3) Add 1ml Trizol and mix well by pipetting;
[0059] (4) Add 200 μl of chloroform, vortex for 20 sec, let stand for 10 min, centrifuge at 12000×rpm for 1 min, and discard the supernatant;
[0060] (5) After centrifuging again at 12000×rpm, discard the supernatant and keep the precipitate, and the white precipitate of DNA at the bottom of the tube can be seen;
[0061] (6) Add 1ml of trisodium citrate to the tube, flick or invert to mix and rinse the precipitate, let stand for 20-30min, centrifuge at 12000×rpm for 5min, and discard the supernatant;
...
Embodiment 2
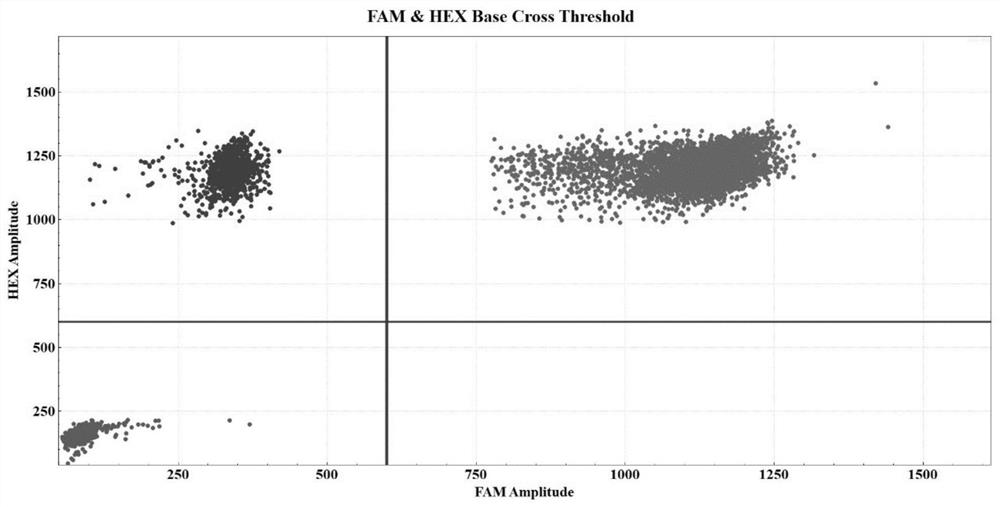
[0084] Take the bone marrow samples of patients with U2AF1 gene Q157R mutation as an example. The specific detection process is the same as that in Embodiment 1, and will not be repeated here. After data analysis, it can be obtained as figure 2 The experimental results and related data.
[0085] In 250ng sample DNA, the FAM concentration is 4712.85copies / μl, the VIC concentration is 4857.83copies / μl, the calculated U2AF1 Q157 mutation template concentration is 144.98copies / μl, and the U2AF1 allele frequency is 2.98%.
Embodiment 3
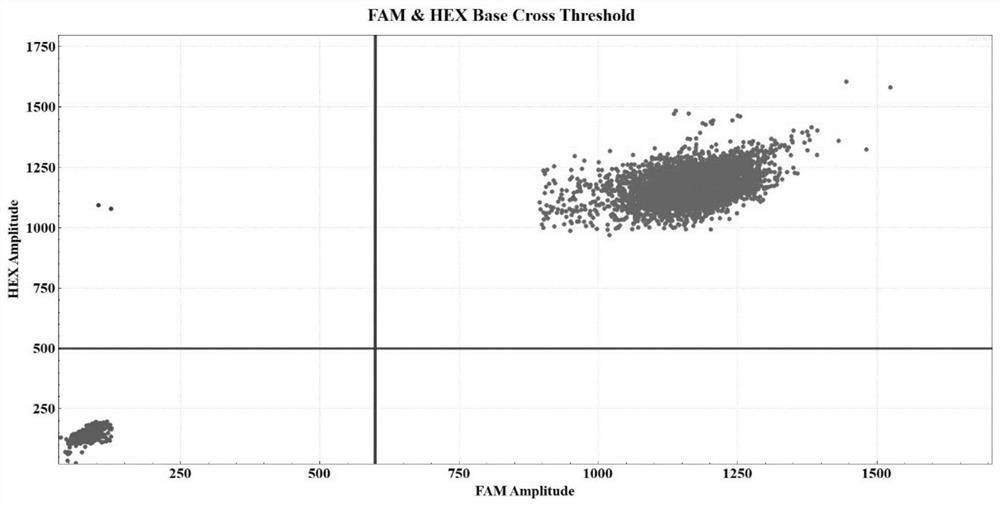
[0087] Take the bone marrow samples of U2AF1 gene Q157 mutation-negative patients as an example. The specific detection process is the same as that in Embodiment 1, and will not be repeated here. After data analysis, it can be obtained as image 3 The experimental results and related data.
[0088] In 250ng sample DNA, the concentration of FAM is 4611.04 copies / μl, the concentration of VIC is 4612.77 copies / μl, the calculated concentration of U2AF1 Q157 mutation template is 1.73 copies / μl, which is lower than the detection limit (8.00 copies / μl), so the patient’s U2AF1 gene Mutation negative.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com