Method and kit for amplifying and detecting nucleic acid
A target nucleic acid and polymerase technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of inability to realize target detection of highly mutated species, influence, and limit popularization and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
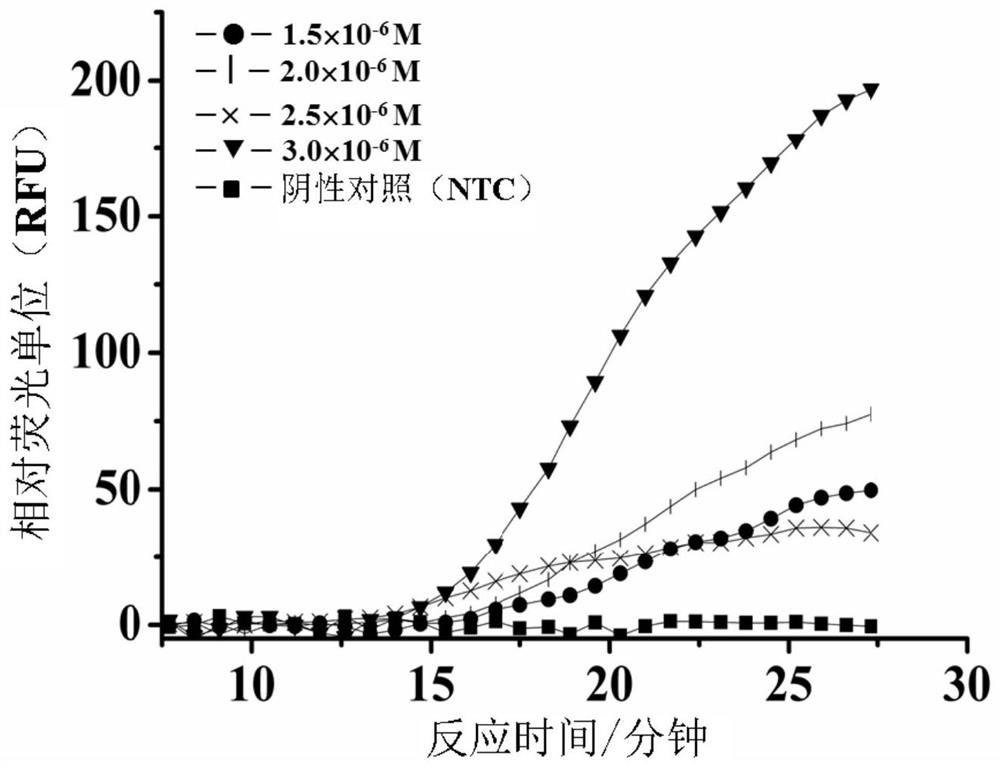
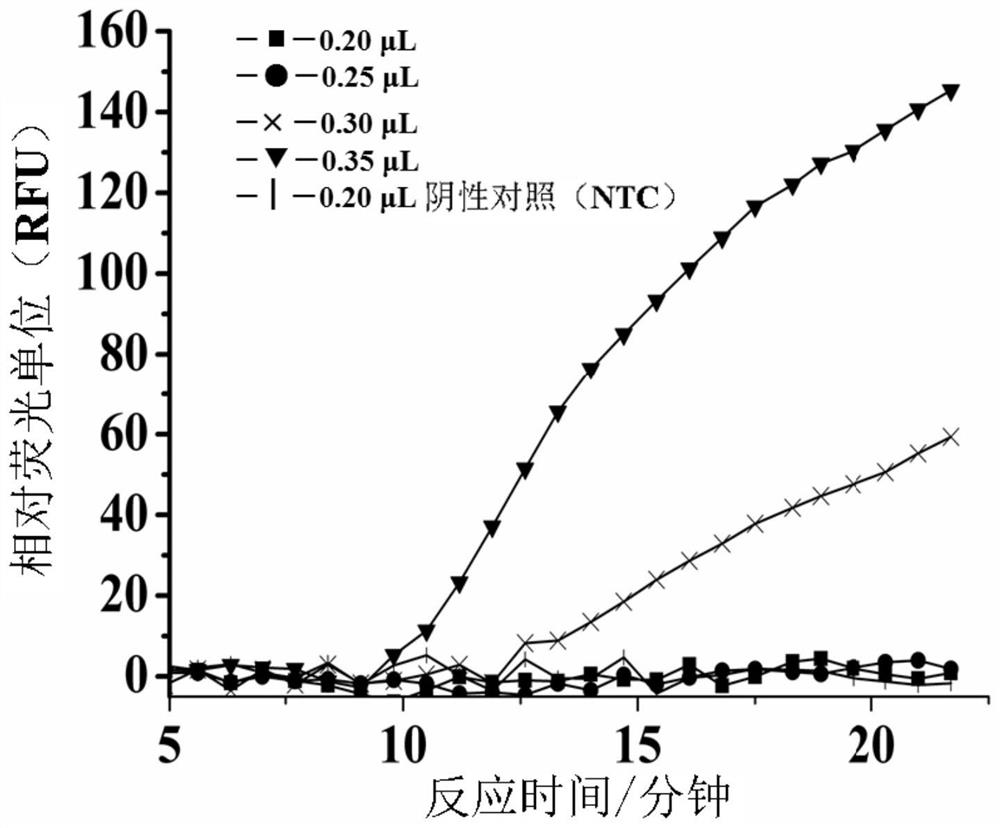
[0220] 5.2 Example 2: Optimization of polymerase concentration for fast strand displacement amplification (SEA) reaction system.
[0221] The following studies were performed to test the effect of polymerase concentration on the rate of fast SEA reactions.
[0222] The same primers (SEQ ID NO: 1 and 2) were designed against the same target sequence in the above L. monocytogenes genome (SEQ ID NO: 1) as described in Example 1 above. Prepare the primers and the L. monocytogenes genome as described above and mix with the other PCR reactions to form a 10 µL amplification mix as shown in Table 2 below. In order to achieve the optimal amplification rate with the polymerase concentration, four amplification mixtures containing different enzyme concentrations were prepared, each containing a final concentration of 3.0×10 -6 M with primers and final concentrations of 0.16 U / μL, 0.20 U / μL, 0.24 U / μL, and 0.28 U / μL (corresponding to 8 U / μL enzyme stock solution of 0.20 μL, 0.25 μL, 0.30...
Embodiment 6
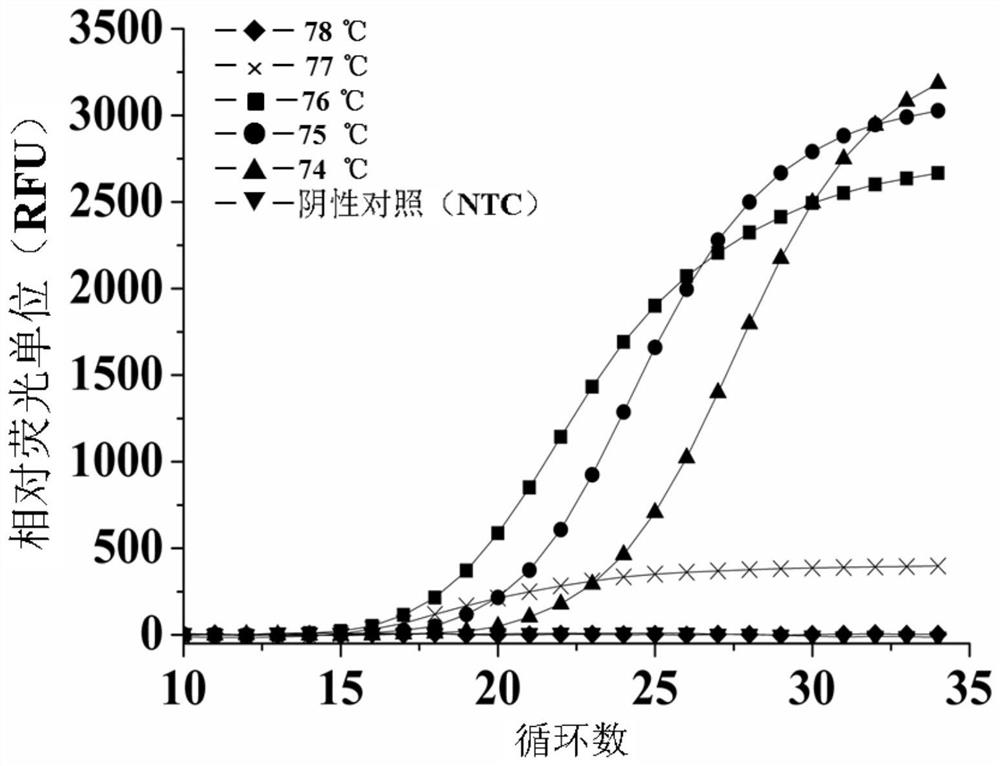
[0255] 5.6 Example 6: Comparison of isothermal SEA reaction under constant temperature conditions and fast SEA reaction under rapid thermal cycle conditions
[0256] The following study compares the isothermal SEA reaction carried out under constant temperature conditions (such as the procedure described in CN 109136337A) and the fast SEA reaction under the current fast thermal cycle conditions.
[0257] Specifically, the same primers (SEQ ID NO: 1 and 2) were designed for the same L. monocytogenes genome (SEQ ID NO: 1) target sequence according to the method described above. The primers and L. monocytogenes genomic material were obtained according to the method mentioned above, and mixed with other PCR reactions to form a 10 μL amplification mixture, the components of which are shown in Table 5 below. In addition, this study set up a series with different initial concentrations (1.0×10 -11 M, 1.0×10 -12 M, 1.0×10 -13 M, 1.0×10 -14 M, 1.0×10 -15 M, 1.0×10 -16 M, 1.0×10 ...
Embodiment 7
[0266] 5.7 Example 7: Primer Design
[0267] Use NUPACK network tool (www.nupack.org), DNAMelt Web (http: / / unafold.rna.albany.edu / ?q=DINAMelt), NOVOPRO www.novopro.cn / tools / rev_comp.html) and NCBI website ( The BLAST algorithm at www.ncbi.nlm.nih.gov / tools / primer-blast) was used for primer design and evaluation.
[0268] DNA primers were synthesized by Personal Biotechnology Co., Ltd. (Shanghai, China). The SEA detection kit was purchased from Navid Biotechnology Co., Ltd. (Qingdao, China). DNA extraction kit was purchased from Tiangen Biotechnology Co., Ltd. (Beijing, China). Other reagents and buffers were of analytical grade.
[0269] traditional PCR reaction : Genomic DNA of Mycoplasma pneumoniae, Chlamydia trachomatis, pork, Bacillus cereus and Staphylococcus aureus were extracted using TIANamp DNA Extraction Kit (Beijing Tiangen Biotechnology Co., Ltd., Beijing) according to the instructions provided by the manufacturer. Using CFX Connect TM Real-time quantitative...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com