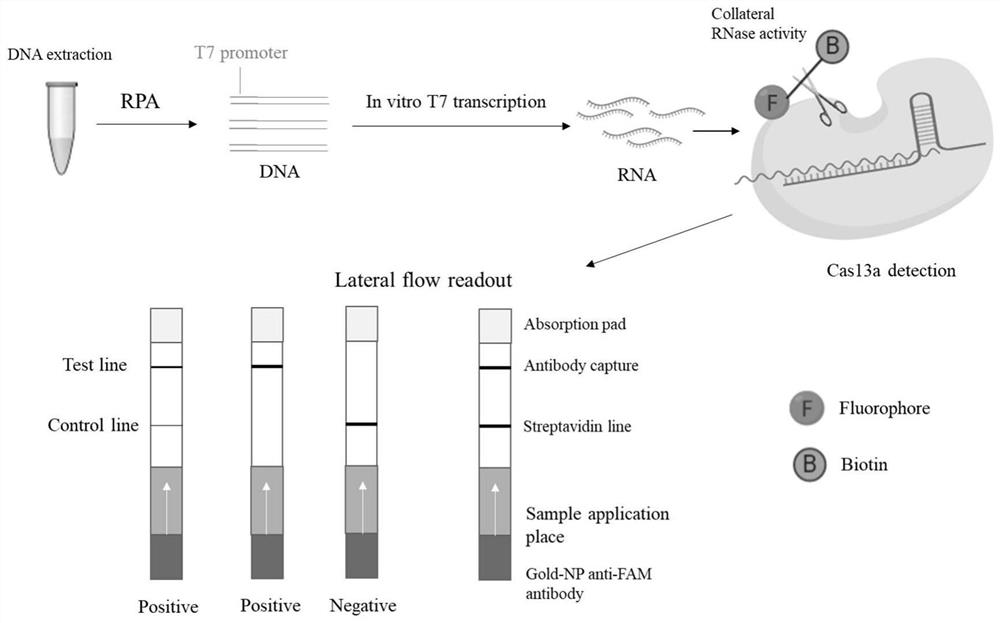
RPA specific primer pair for detecting ALV-A/B/J, crRNA fragment and application thereof
A primer pair and specificity technology, applied in the field of RPA-specific primer pair detection of ALV-A/B/J, can solve the problems of unsuitable field detection, expensive equipment, and high technical requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Establishment of the CRISPR-Cas13a system for detecting ALV-A / B / J
[0052] 1. Design of RPA primers and crRNA
[0053] Retrieve the nucleotide sequence of the gp85 gene of ALV-A / B / J from the Genbank database of NCBI, and select a section that is relatively conserved in the gp85 gene of the same subgroup strain and is different from other subgroup ALV strains through sequence alignment and screening. Nucleic acid sequences with large differences are used as target regions. According to the method recommended in the TwistDx instruction manual of the RPA kit seller, several pairs of primers were designed for ALV-A / B / J (Table 1). A 25bp T7 promoter sequence (underlined part of Table 1) was added to the 5' end of the upstream primer, so that the target fragment amplified by RPA could be transcribed as the recognition object RNA of Cas13a. The size of the target fragment obtained after RPA amplification is about 140-190bp, and a 28bp highly conserved crRNA targeti...
Embodiment 2
[0080] Example 2 The specificity and sensitivity of the method for detecting ALV-A / B / J based on CRISPR-Cas13a
[0081] 1. Specificity
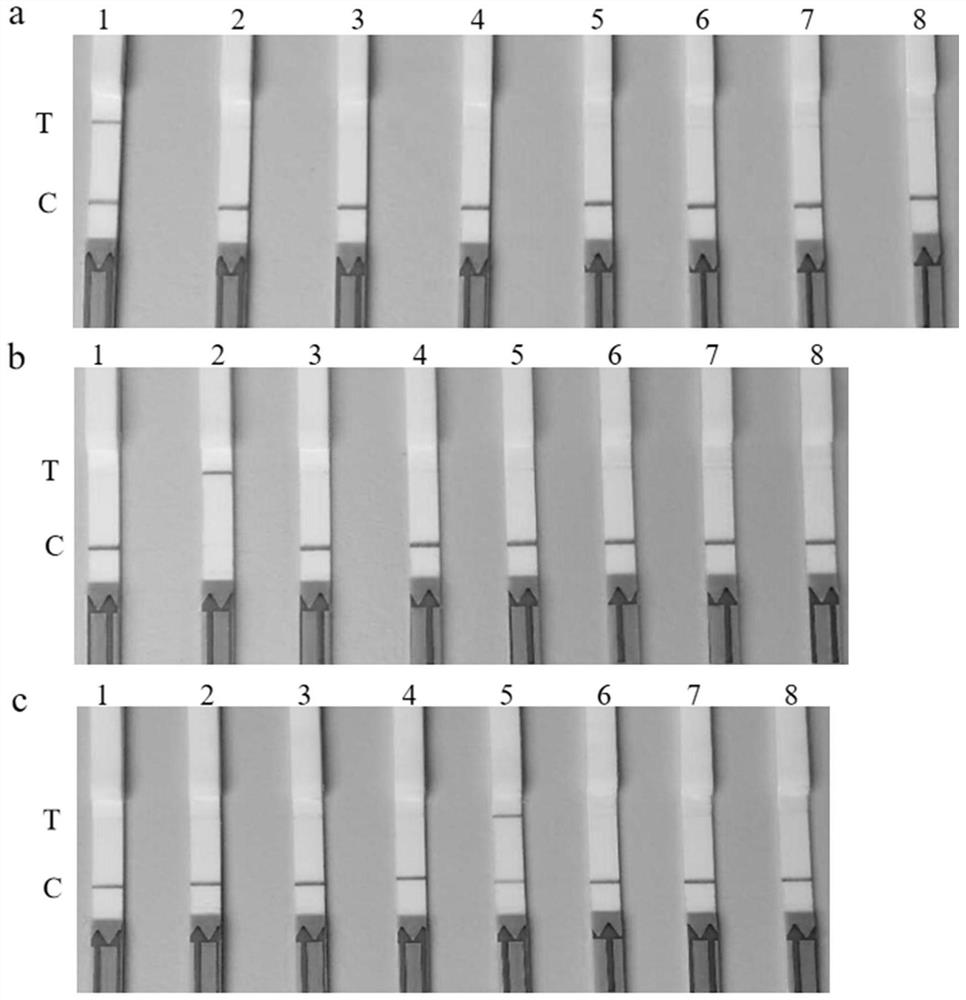
[0082] The proviral DNA of other ALV subgroup strains (C, D, E) infecting chickens, the proviral DNA of avian reticuloendotheliosis virus (REV) and the DNA of chicken Marek's virus (MDV) were used as templates, respectively , configure the RPA and Cas13a reaction system according to the method in Example 1 to verify the specificity of the present invention. like image 3 As shown, only ALV-A / B / J has a "T" line that is positive; other nucleic acid tests have only a "C" line that is negative. The results show that the invention has high specificity and no cross-reaction with other chicken tumor viruses.
[0083] 2. Sensitivity
[0084]Use the specific identification primers (H5 / envA, H5 / envB, H5 / H7) of ALV-A / B / J in Table 3 to amplify the corresponding sequences and clone them into the TA vector GEM-T-easy. The recombinant plasmid was used a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com