A method for improving enzymes
A mutant and protein technology, applied in the fields of molecular biology and protein engineering, can solve problems such as limited effects, achieve the effects of improving efficiency, improving reliability and screening efficiency, improving protein stability and protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
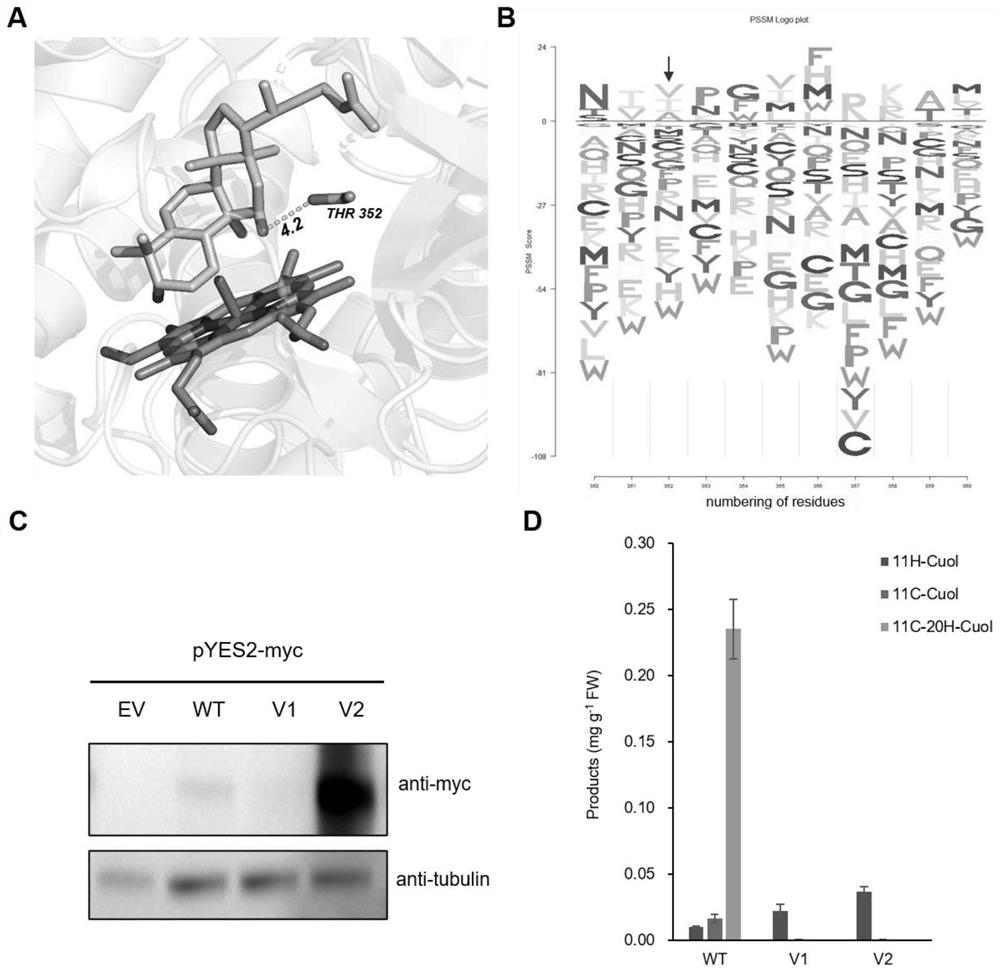
[0063] Example 1. Carrying out structural simulation of proteins and obtaining high-precision protein structure models
[0064] Structural modeling of CPY87D20 (cucumber P450 enzyme) was performed using the homology modeling function of Rosetta. First, the fasta amino acid sequence file of CPY87D20 was used as input, and the homologous sequence of CPY87D20 was searched from the pfamA_32.0 or scope70_1_1.75 database using hhblits to obtain a multiple sequence alignment file in a3m format. Take this file as input, use hhsearch to retrieve the existing pdb structure from the pdb70 database, and obtain the pdb.hhr file. Use this file as input for homology modeling with RosettaCM. At the same time, the sdf structures of the ligands cucurbitenol (Cuol) and heme (heme) were obtained from Pubchem, and the parameter files were generated using the Rosetta program. Use Rosetta Ligand_Docking to dock heme and Cuol into the protein model to obtain the complex. Using the deep learning fr...
Embodiment 2
[0065] Embodiment 2, change the catalytic activity of enzyme
[0066] Using the psiblast program and using uniref90 as the database, the position-specific scoring matrix PSSM of CPY87D20 was obtained, which displayed the conservation information of each amino acid of CPY87D20. The PSSM scoring matrix is analyzed to obtain potential replacements of catalytic centers or protein surface amino acids according to the purpose of the experiment (improving activity or stability). Using hhblits and uniclust30_2018_08 as the database, multiple sequence alignments were performed on CPY87D20, amino acid co-evolution analysis was performed using Gremlin, and potential replacement sites were obtained according to the purpose of the experiment. Selection of Substrate Cucurbitadienol Based on Protein Model Amino acids within the range exclude amino acid sites near the heme (because it may cause loss of activity). Replacement mutations with PSSM scores greater than 0 were screened against...
Embodiment 3
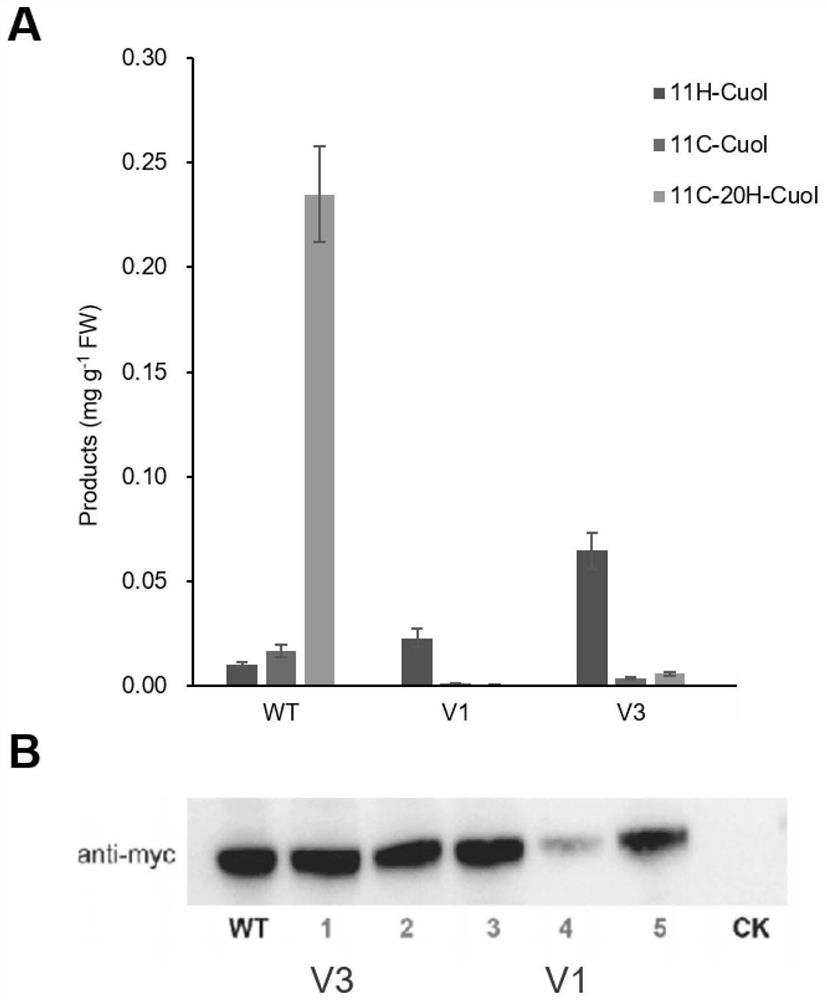
[0072] Embodiment 3, the effect of each mutant
[0073] The P450s mutant of the present invention is based on the amino acid sequence of CPY87D20 of cucumber (cucumis sativus), as shown in SEQ ID No.1, and multiple mutants including V4 have been obtained successively, for example: mutant V1, amino acid sequence As shown in SEQ ID No.2; mutant V2, the amino acid sequence is shown in SEQ ID No.3; mutant V3, the amino acid sequence is shown in SEQ ID No.4.
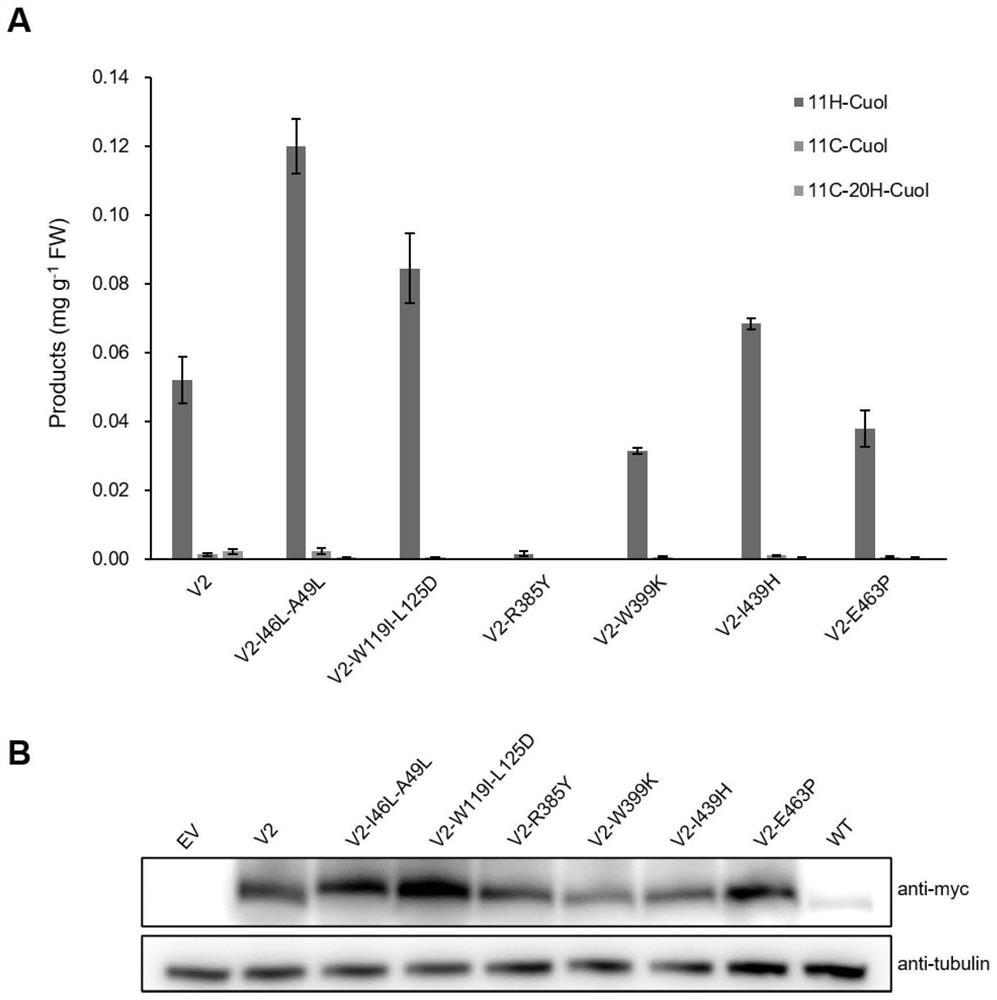
[0074]For another example, mutants further mutated on the basis of the above mutants: V2-I46L-A49L, V2-W119I-L125D, V2-R385Y, V2-W399K, V2-I439H, V2-E463P, V2-I46L-A49L- C343Y, V3-C343Y, V3-C343Y-S49L, V3-C343Y-I46L, V3-K73Y, V3-F89D, V3-Y432E, V3-L125D, V3-R383T, V3-W399D, etc., tested in batches under the same conditions . Several were found: both the yield of the target product 11H-Cuol, the substrate selection specificity of the enzyme, and the protein expression level were significantly higher than those of the wild ty...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com