Olivol synthase variants and engineered microorganisms expressing them
An olivetolic acid cyclase and engineering technology, applied in the field of microorganisms and enzymes, can solve the problems of difficult to obtain high-purity products, harsh conditions, limited production capacity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
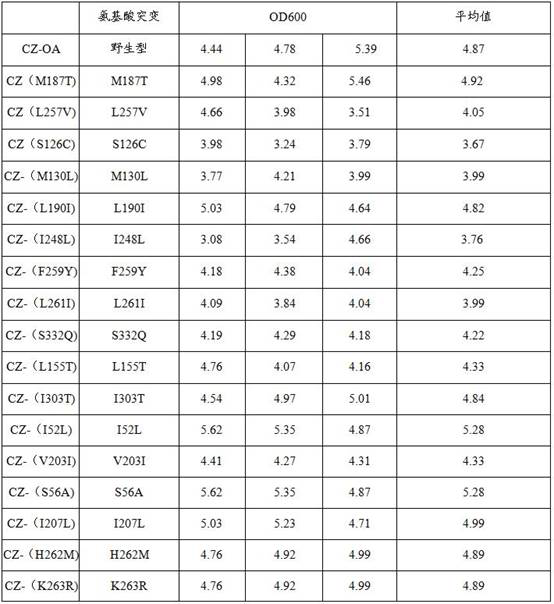
[0067] Item 1. An engineered Escherichia coli, wherein the engineered Escherichia coli is modified to express an olivetol synthase (OLS) variant, wherein the OLS variant comprises one of the following mutations compared to its wild type One or more: I303T, I52L, S56A, H262M and K263R.
[0068] Item 2. The engineered E. coli according to any one of the preceding items, wherein the engineered E. coli is modified to express olivate cyclase (OAC).
[0069] Item 3. The engineered Escherichia coli according to any one of the preceding items, wherein the genomic fabH gene of the engineered Escherichia coli is deleted.
[0070] Item 4. The engineered Escherichia coli according to any one of the preceding items, wherein the genome fadE gene of the engineered Escherichia coli is deleted.
[0071] Item 5. The engineered E. coli according to any one of the preceding items, wherein both the fabH and fadE genes of the engineered E. coli genome are deleted.
[0072] Item 6. The engineered ...
Embodiment 1
[0101] Example 1: Deletion of the fabH gene
[0102] The fabH gene in the genome of Escherichia coli BW25113 was deleted to reduce the flow of intracellular Malonyl-CoA to the branch metabolism, thereby increasing the accumulation of intracellular Malonyl-CoA to further increase the synthesis of the target product OA.
[0103] Design and synthesize the H1-kana-H2 described in SEQ ID NO: 1, according to the literature (Datsenko K A, Wanner BL. One-step inactivation of chromosome genes in Escherichia coli K-12 usingPCR products[J]. Proceedings of the National Academy of Sciences, 2000, 97 (12): 6640-6645.) provided a method to integrate SEQ ID NO: 1 into the fabH gene position of the genome of BW25113 through λ-Red homologous recombination to delete the fabH gene, the steps are as follows:
[0104] 1. Prepare the competent state of BW25113;
[0105] 2. Introduce plasmid pKD46;
[0106] 3. Inoculate BW25113 (pKD46) into 3 mL of LB containing ampicillin, the concentration of a...
Embodiment 2
[0126] Example 2: Deletion of the fadE gene
[0127] In the engineered BW25113 obtained in Example 1, the fadE gene in the genome was further deleted to reduce the flow of intracellular hexanoyl-CoA to the bypass metabolism, thereby increasing the accumulation of intracellular hexanoyl-CoA to further increase the synthesis of the target product OA .
[0128] Design and synthesize the H3-kana-H4 described in SEQ ID NO: 4, as described in Example 1, integrate SEQ ID NO: 4 into the fadE gene position of the genome of BW25113 that lacks the fabH gene so that the fadE gene is deleted and integrated SEQ ID NO: 4 IDNO: 4 to delete the KanR resistance gene.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com