Use of ligands derived from receptor-binding domain of porcine endogenous retrovirus type b for diagnosing SMVT-related diseases
A technology for infectious diseases and ligands used in the field of measuring SMVT levels in biological samples, which can solve problems such as restricting development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
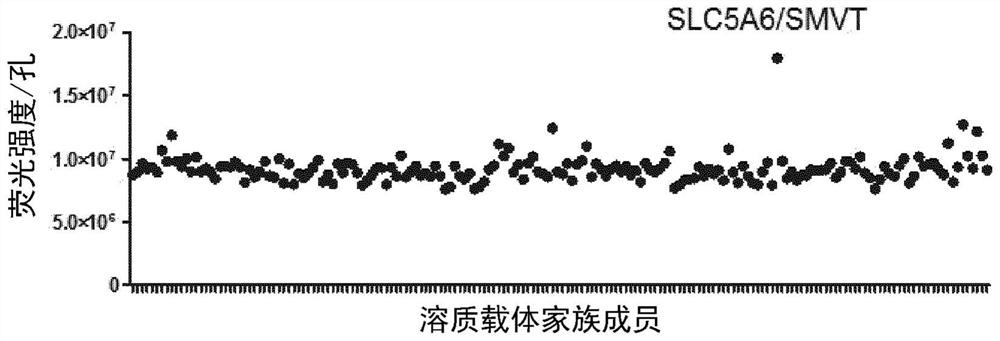
[0350] Example 1: Automated screening for PERVB-RBD identifies SLC5A6 as a PERVB-RBD cognate receptor
[0351] Materials and Methods
[0352] Three independent screens were performed on PERV-B-RBD (SEQ ID NO: 13) binding of a total of 172 solute carrier protein (SLC) family members using the Robot, images and fluorescence intensity parameters were acquired by Cellomics (ArrayScan XTI HCS Thermo Scientific).
[0353] 20,000 quail QT6 cells per well were seeded in 96-well plates coated with poly-D-lysine. After 24 hours, cells were transfected with 100 ng of SLC expression vector using JetPrime transfection reagent (Polypus Transfection 114-15) according to the manufacturer's instructions. QT6 cells were chosen because the spontaneous binding of PERV-B.RBD ligand was the lowest of the cell lines considered and tested. The next day cells were washed with PBS and incubated in fresh DMEM / FBS for 48 hours prior to binding assay. Binding assays were performed on transfected ad...
Embodiment 2
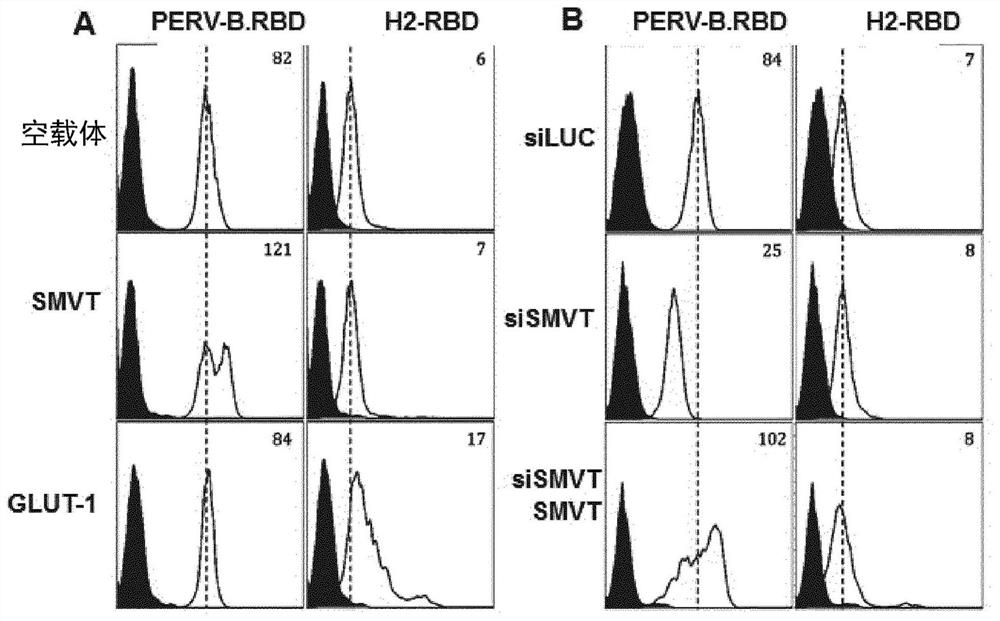
[0356] Example 2: Binding specificity of PERV-B.RBD ligands to SMVT / SLC5A6
[0357] Materials and Methods
[0358] Will 5x10 5 HEK293T cells were seeded on 6-well plates and transfected with siRNA against firefly luciferase gene (siLUC, 5'-CUUACGCUGAGUACUUCGA-3'-SEQ ID NO: 15), or siRNA against SLC5A6 gene (siSMVT, 5'-GGAUGAGUCUUGGUGUGUUTT-3'-SEQ ID NO: 16). Forty-eight hours after transfection, cells were deplated with PBS containing 1 mM EDTA. For binding assays, 1x10 5 3 cells were resuspended in 100 μl of PBA (PBS with 2% FBS) containing saturating concentrations of PERV-B.RBD (SEQ ID NO: 13) or human T-cell leukemia type 2 known to bind SLC2A1 / GLUT1 Virus (HTLV2)-RBD and incubated at 37°C for 30 minutes. Cells were then washed twice with 100 μl of PBA, incubated with Alexa488-conjugated anti-mouse IgG1 for 20 min at 4°C, washed twice and resuspended in 200 μl of PBA. Results were acquired by flow cytometry on a FACSCalibur (Becton Dickinson) and analyzed by FlowJo...
Embodiment 3
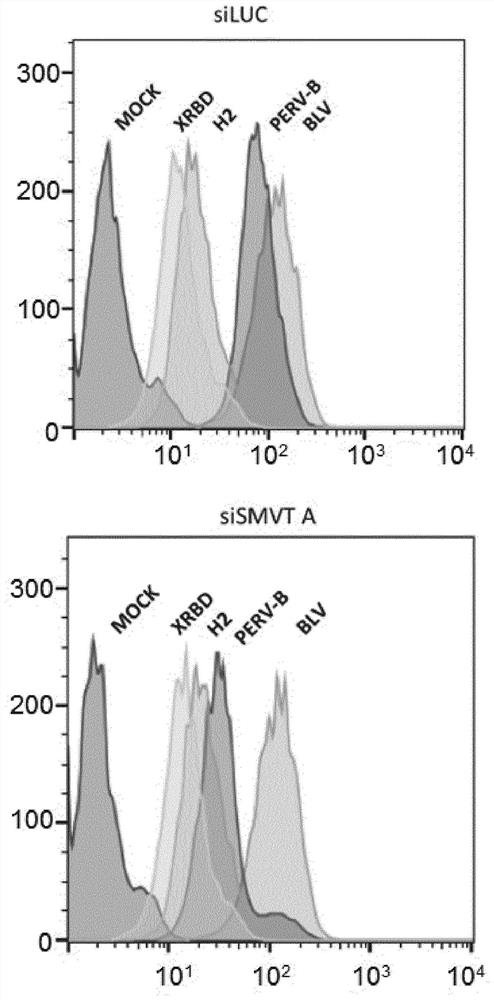
[0361] Example 3: Binding specificity of PERV-B.RBD ligands to SMVT / SLC5A6, assessed using secondary anti-SMVT siRNA
[0362] Materials and Methods
[0363] Will 5x10 5 HEK293T cells were seeded on 6-well plates and transfected with: siRNA against the firefly luciferase gene (siLUC, 5'-CUUACGCUGAGUACUUCGA-3'-SEQ ID NO: 15), or against the SLC5A6 gene and different from the example siRNA of the siRNA used in 2 (siSMVT A, 5'GCAGGAUCAUGCCAGAAAUTT-3'-SEQ ID NO: 23). Forty-eight hours after transfection, cells were deplated with PBS containing 1 mM EDTA. For binding assays, 1x10 5 Each cell was resuspended in 100 μl of PBA (PBS containing 2% FBS) containing a predetermined saturating concentration of PERV-B.RBD (SEQ ID NO: 13) or RBD from a heterophile murine retrovirus (XRBD, which receptor known as XPR1 / SLC53A1) or HTLV2-RBD (H2, whose receptor is known as GLUT1 / SLC2A1) or RBD derived from bovine leukemia virus (BLV) (whose receptor is known as CAT1 / SLC7A1) or not Control ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com