Method for high-throughput identification of interaction between pathogenic bacteria and host molecules
A pathogenic bacteria, high-throughput technology, applied in the field of pathogenic bacteria identification, can solve the problems of low screening efficiency and operational errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] Next, the technical solutions in the embodiments of the present invention will be described in connection with the drawings of the embodiments of the present invention, and it is understood that the described embodiments are merely the embodiments of the present invention, not all of the embodiments. Based on the embodiments of the present invention, all other embodiments obtained by those of ordinary skill in the art are in the range of the present invention without making creative labor premise.
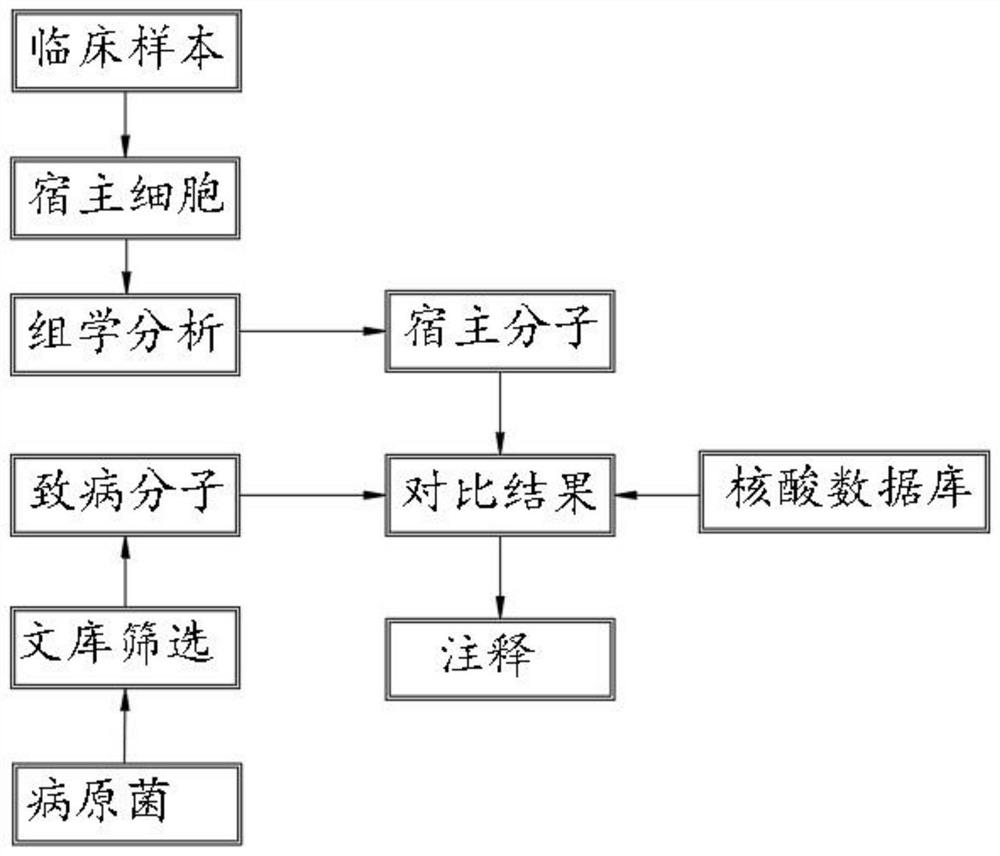
[0021] according to figure 1 Looking:
[0022] S1, select 100 tuberculosis H37RV toxic associated proteins to construct the bait carrier, screening and literature excavation tuberculosis H37RV with host interaction proteins, identifying three known nominal bacillus H37RV intrusion and survival related known The host cell key receptor and key signal transduction molecule screens 40 new candidate host cytokines associated with NOTE M. octrue H37RV, which is built into a transposon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com