Vector system for selection of genes encoding secreted proteins and membrane-bound proteins
a gene selection and membrane-bound protein technology, applied in the field of vector system for selecting genes encoding secreted proteins and membrane-bound proteins, can solve the problems of low transfection efficiency, signal sequence trapping using mammalian cells, and difficult identification of secretion signal sequences that are normally present in the 5′ end of cdna encoding, so as to improve the overall efficiency of cloning cdnas
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Amplification of cDNAs Encoding Secreted or Membrane-Bound Proteins Containing Specific Amino Acid Motifs
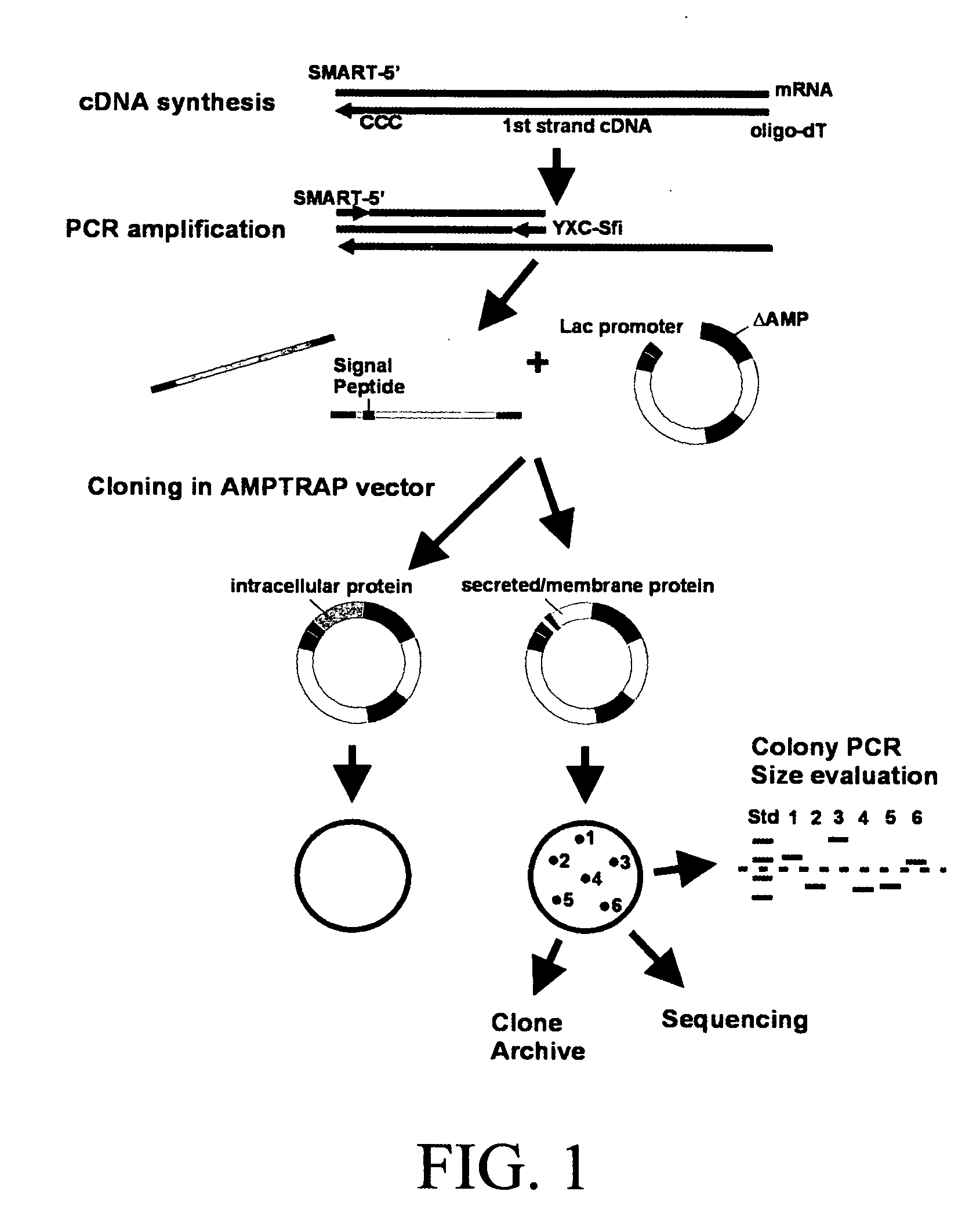
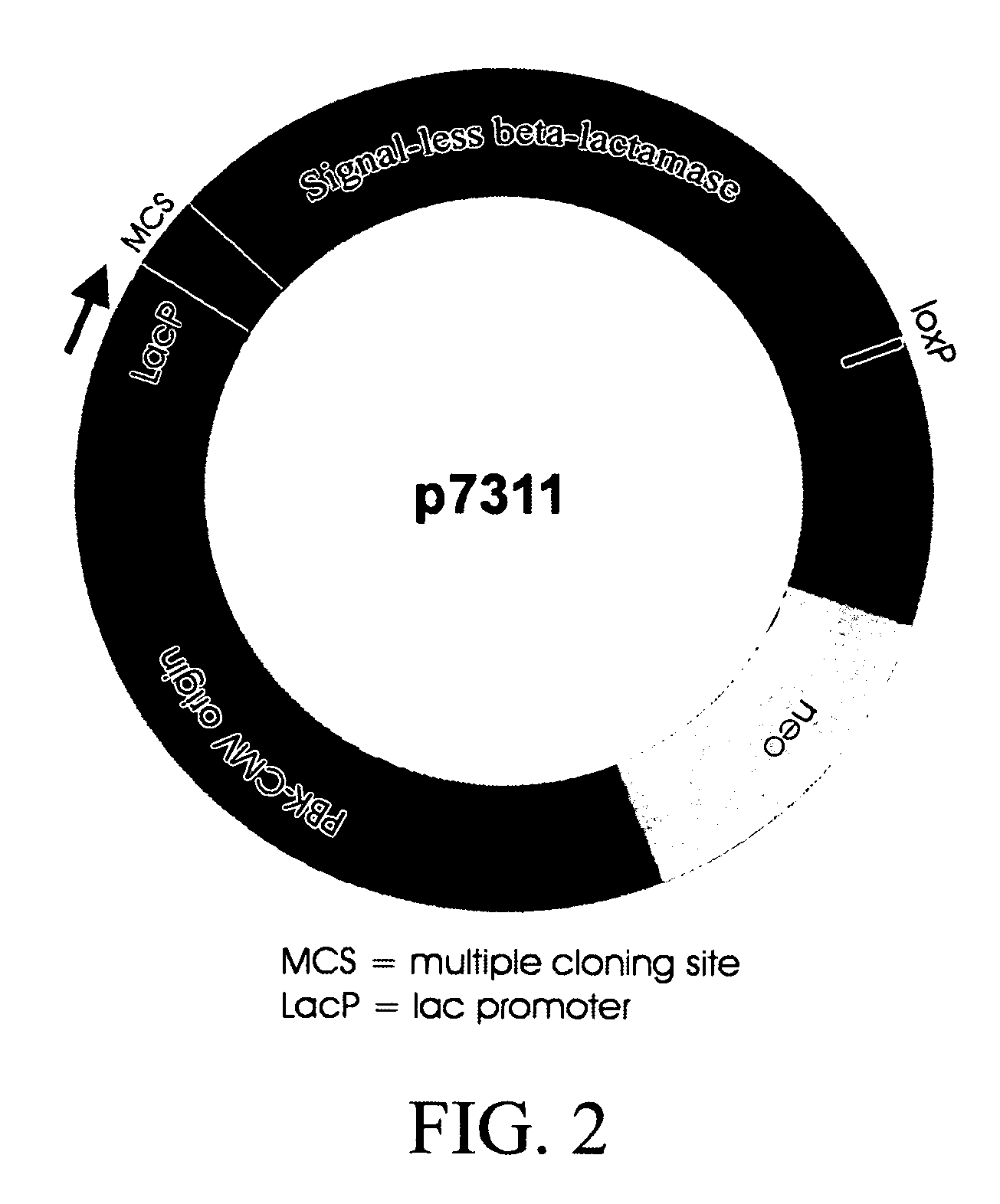
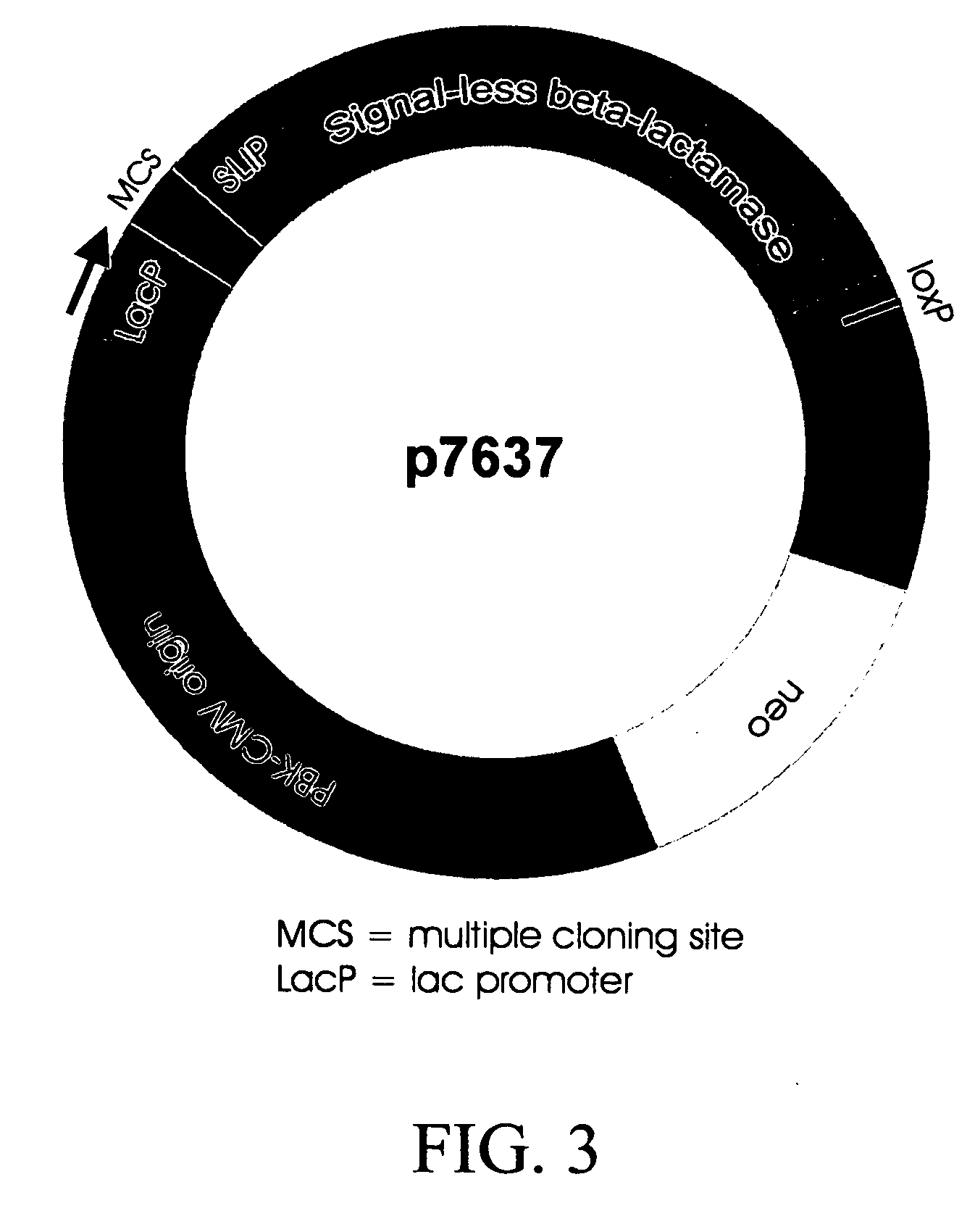
[0066] In order to select cDNAs that encode secreted or membrane-bound proteins, the cloning vector G7311 (referred to herein as an “Amptrap” vector) was designed by the present inventors, which allows selection for signal sequence-encoding regions by β-lactamase rescue (as shown in FIG. 2). The Amptrap vector shown contains a 5′-truncated β-lactamase gene driven by a Lac promoter. The product of the modified β-lactamase gene lacks the signal peptide present in the wild-type protein and is therefore unable to be exported into the bacterial periplasmic space. An asymmetric pair of SfiI sites for insertion of cDNA lies immediately 5′ to the β-lactamase open reading frame (ORF). A separate neomycin phosphotransferase marker was included to allow propagation of the vector without cDNA inserts. Because β-lactamase must be secreted into the periplasmic space to produce ampicillin resi...
example 2
Cloning of Expressed Sequence Tags (EST) Sequences Using the Amptrap Vector
[0068] cDNA was synthesized from Florida lancelet (Branchiostoma floridae) and sea lamprey (Petromyzon marinus) tissues and cloned into an Amptrap vector. 57 sequences were analyzed by BLASTX searching of the Genbank database. Although 17 sequences failed to match known sequences in Genbank, all of the remaining 40 sequences were found to encode proteins that are likely to be secreted or bound to membranes (Table 1).
TABLE 1Beta-lactamase fusion (“amptrap”) EST sequencesG#TissueSourceStock?Gel dateComments7024PMP20000827, clone 3Y20000928no match*7026PMI20000827, clone 5Y20000830trypsinogen b1†7027PMI20000827, clone 6Y20000830no match*7031PMI20000827, clone 10Y20000830trypsinogen b1†7033PMI20000827, clone 12Y20000830trypsinogen b1†7034PMI20000827, clone 13Y20000905trypsinogen b1†7035PMI20000827, clone 14Y20000905chymotrypsinogen†7037PMI20000827, clone 16Y20000905trypsinogen†7038PMI20000827, clone 17Y2000090...
example 3
Amplification of Candidate Immune-Type Receptor Genes from Branchiostoma floridae, Raja eglanteria, and Petromyzon marinus
[0069] In order to identify potential new members of the novel immune-type receptor (NITR) gene family previously described in teleost fish, cDNA sequences from Florida lancelet Branchiostoma floridae, clearnose skate Raja eglanteria, and sea lamprey Petromyzon marinus tissues were amplified by 5′-RACE PCR using various 3′ primers and the 5′-SMART oligonucleotide. These primers included:
(SEQ ID NO. 8)nitrVYWFR-5′TGGCCGAGGCGGCCCNCGRAACCARTANAC-3′;Sfi:(SEQ ID NO. 9)nitrVYWF-Sfi:5′GACTGGCCGAGGCGGCCCRAACCARTANAC-3′;(SEQ ID NO. 10)nitrYWFR-Sfi:5′-GACTGGCCGAGGCGGCCCNCGRAACCARTA-3′;(SEQ ID NO. 11)nitrYWFK-Sfi:5′-GACTGGCCGAGGCGGCCCYTTRAACCARTA-3′;(SEQ ID NO. 12)nitrWFR1-Sfi:5′-GACTGGCCGAGGCGGCCCNCGRAACCA-3′;(SEQ ID NO. 13)nitrWFR2-Sfi:5′-GACTGGCCGAGGCGGCCCYCTRAACCA-3′;and(SEQ ID NO. 14)nitrWFK-Sfi:5′GACTGGCCGAGGCGGCCCYTTRAACCA-3′.
[0070] The pool of amplicons were subs...
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid | aaaaa | aaaaa |
| nucleic acid sequence | aaaaa | aaaaa |
| nucleic acid sequences | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com