SNP marker for identifying apricot peel hair traits, primer and application
A technology for identifying and marking apricots, which is applied in the field of molecular biology, can solve the problems of long cycle of target varieties, achieve low cost, accelerate the breeding process, and shorten the breeding period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The following examples are used to illustrate the present invention, but are not intended to limit the scope of the present invention. Unless otherwise specified, the examples are all in accordance with conventional experimental conditions, such as Sambrook et al. Molecular Cloning Experiment Manual (Sambrook J & Russell DW, Molecular Cloning: a Laboratory Manual, 2001), or in accordance with the conditions suggested by the manufacturer's instructions. Example 1 Using 46 Li Guangxing fruit offspring to verify the hairy / glabrous traits of the fruit epidermis
[0050] Implementation time: 2020;
[0051]Implementation site: Beijing Forestry and Fruit Tree Science Research Institute;
Embodiment approach
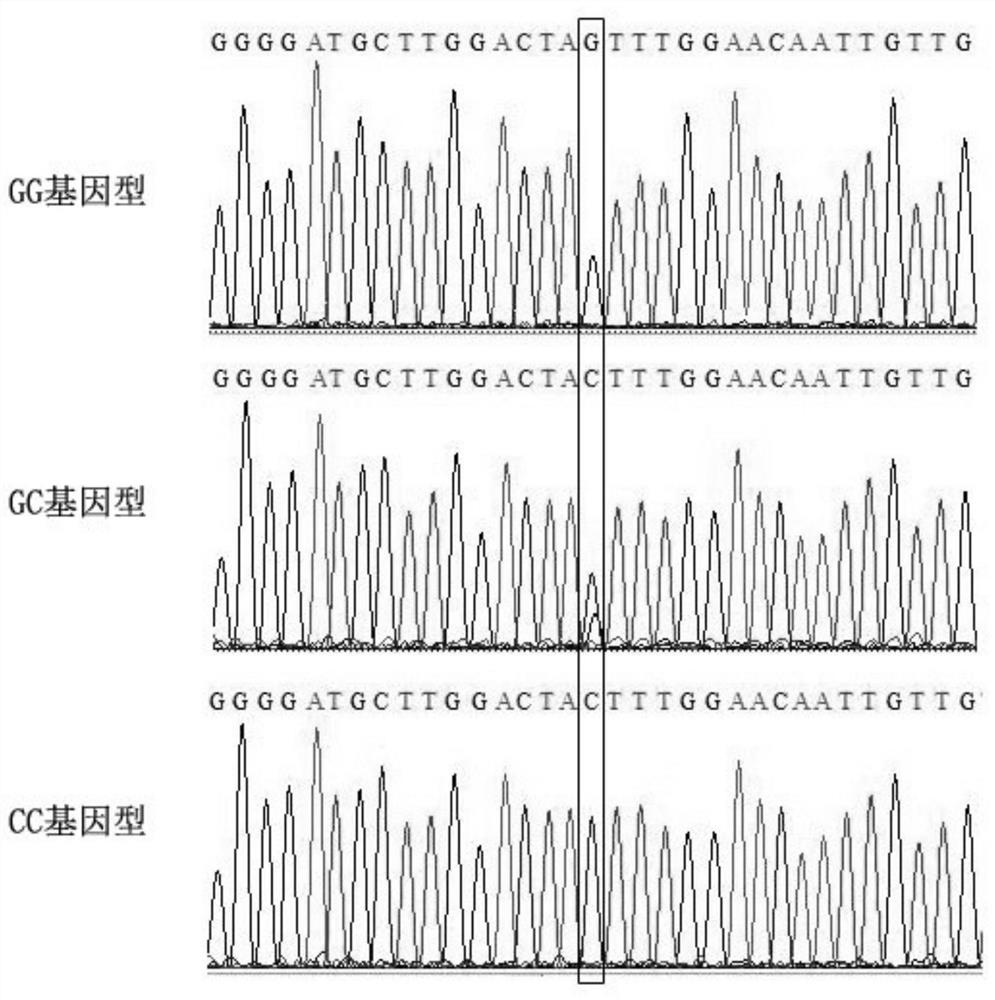
[0053] The population of seed progeny of Li Guangxing was selected, which was a half-sib family. The total DNA of 46 leaves of seedlings of Li Guangxing was extracted by CTAB method, and the purity of DNA was identified. Then use primers (SEQ ID NO:2-3), and carry out PCR reaction according to the above-mentioned steps, after gel electrophoresis of the obtained PCR amplification product, use common agarose gel DA recovery reagent to recover the target fragment, and carry out a generation Sequencing was performed to detect the genotype of the polymorphic site corresponding to the SNP marker of the PCR product, and the results are shown in Table 3.
[0054] Table 3 Verification results of hairy / glabrous epidermis of 46 Li Guangxing apricot offspring
[0055]
Embodiment 2
[0056] Example 2 Select 244 apricot germplasm resources to verify the hairy / hairless traits of the fruit epidermis
[0057] Implementation time: 2020;
[0058] Implementation site: Beijing Forestry and Fruit Tree Science Research Institute;
[0059] implementation plan:
[0060] (1) Total DNA was extracted from leaves of 244 apricot germplasm resources by CTAB method, and the purity of DNA was identified.
[0061] (2) Synthesize primers (SEQ ID NO:2-3), and carry out PCR reaction according to the above steps to obtain PCR amplification products. Sequencing, and then detecting the genotype of the polymorphic site corresponding to the SNP marker of the PCR product.
[0062] (3) Detection of the correlation between SNP molecular markers and hairy and hairless traits
[0063] Use the above primers to verify the reliability of 244 pairs of SNP molecular markers. The genotypes GG and GC are hairy in the apricot fruit epidermis, and CC is hairless. The fruit epidermis of all apri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com