Compositions and methods for improved gene editing
A gene and editing enzyme technology, applied in the field of compositions and methods for improved gene editing, can solve the problems of genome inconsistency, low base editing efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0329] Example 1 experimental scheme
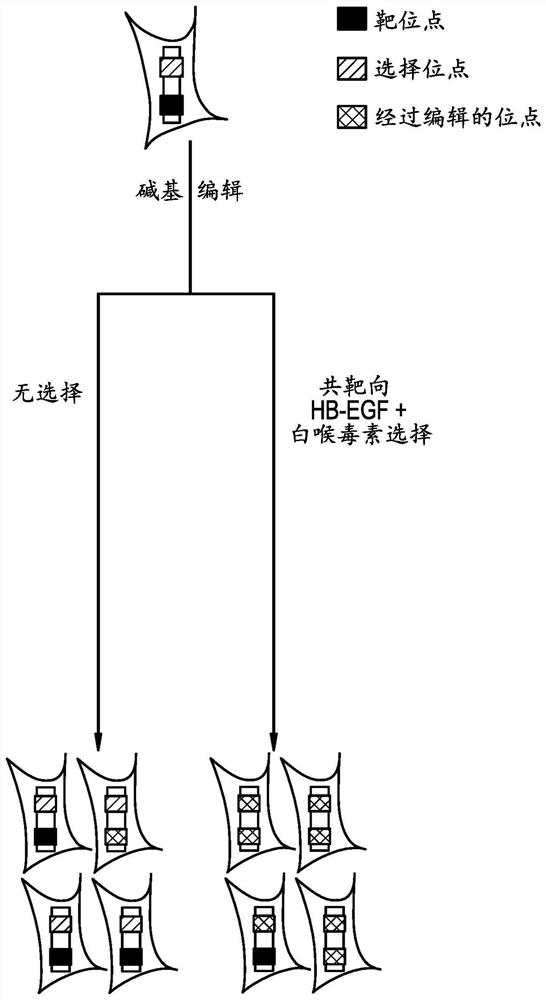
[0330] In this example, a protocol for co-targeted enrichment is provided.
[0331] Cell lines expressing heparin-binding EGF receptors were maintained in culture and subcultured every 2-3 days until transfection. Cells should be >80% confluent on the day of transfection.
[0332] Cells are transfected with a plasmid encoding a base editor or Cas9, and / or together with a plasmid encoding a guide RNA targeting HB-EGF and the gene of interest. Prepare DNA-lipid complexes for transfection according to the manufacturer's protocol. Alternatively, mRNA and RNP complexes can also be used.
[0333] Complexes were added to plates seeded with fresh trypsinized cells the day before.
[0334] The medium was removed 72 hours after transfection, and the cells were trypsinized and replated into new plates with twice the surface area of the previous plate.
[0335] The next day, diphtheria toxin was added to the wells at a concentration of 20 ng / m...
example 2
[0338] Example 2. Screening of guide RNA
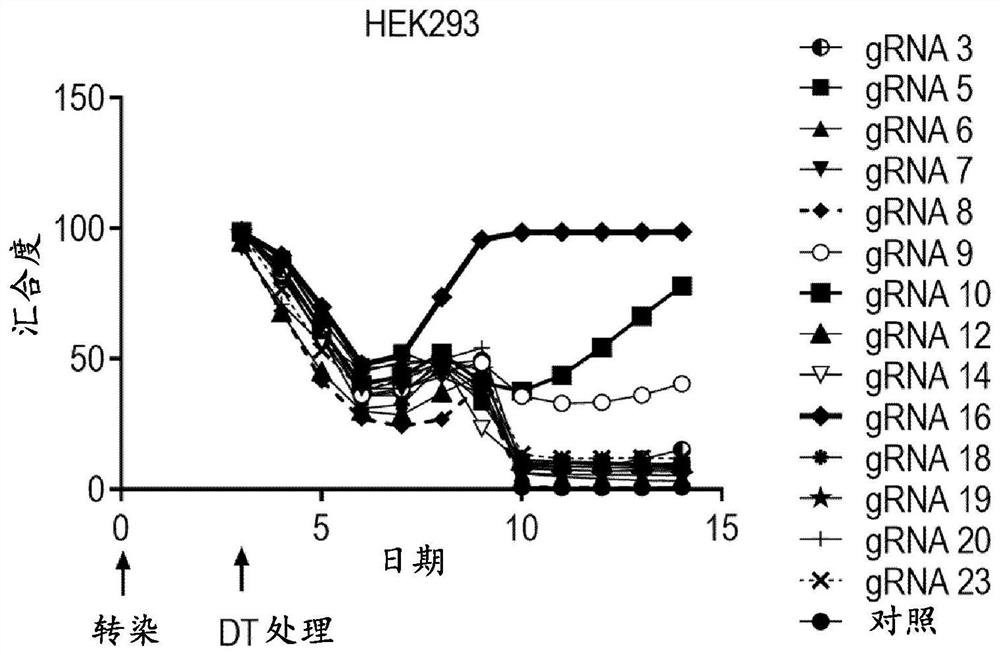
[0339] In this example, guide RNAs (gRNAs) were screened to identify gRNAs that would confer resistance to diphtheria toxin when co-transfected with BE3. Design a panel of gRNAs to tile through the EGF-like domain of HB-EGF (see Figure 4C ). Each gRNA was co-transfected into HEK293 or HCT116 cells with BE3 at a transfection weight ratio of 1:4.
[0340] Cells were treated with 20 ng / mL diphtheria toxin on day 3 post-transfection and then again on day 5 post-transfection. Cell growth was measured by confluency using an INCUCYTE ZOOM.
[0341] Figure 4A with Figure 4B The results shown show that HEK293 and HCT116 cells co-transfected with HB-EGF gRNA 16 and BE3 had the highest growth level among all transfected cells, respectively. Figures 5B-5D The results of sanger sequencing and next-generation sequencing analyzes shown revealed that resistance to diphtheria toxin in gRNA 16-transfected cells was the result of the E141K mut...
example 3
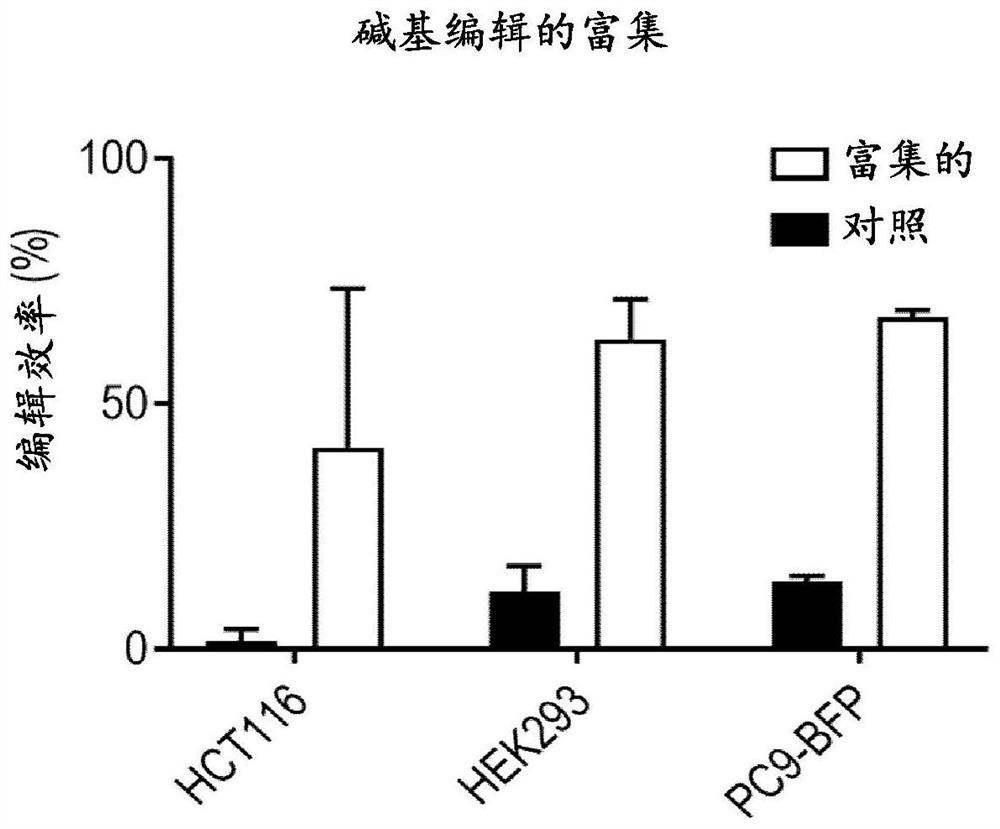
[0342] Example 3. Co-targeted enrichment with BE3 and Cas9
[0343] In this example, co-targeted enrichment using diphtheria toxin selection was tested using BE3 and Cas9, targeting gRNAs identified in Example 2 and gRNA 16 were co-transfected to generate diphtheria toxin resistant cells.
[0344] Plasmid construction
[0345] Cas9 plasmid: DNA sequences encoding SpCas9, T2A self-cleaving peptide, and puromycin N-acetyltransferase were synthesized by GeneArt (GeneArt) and cloned into expression vectors with CMV promoter and BGH poly-A tail. see Figure 15 The plasmid map of .
[0346] BE3 plasmid. The DNA sequence of base editor 3 was synthesized by Genentech AG using the restriction sites BamHI and XhoI and cloned into pcDNA3.1(+). see Figure 14 The plasmid map of .
[0347] gRNA plasmid. A complementary primer pair (5'-AAAC-N20-3' and 5'-ACCG-N20-3') was used to introduce the target sequence of the gRNA at the AarI cleavage site in the template plasmid. The templa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com