Method for detecting polynucleotide kinase in high-salt high-protein biological sample
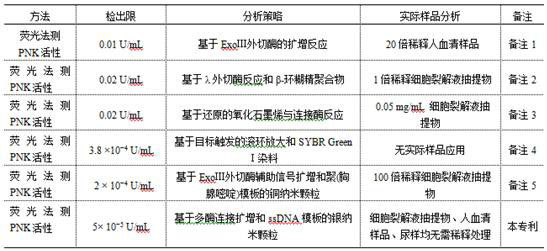
A nucleotide kinase and high protein technology, which is applied in the field of detecting polynucleotide kinases in high-salt and high-protein biological samples, can solve the problems of complex operation and inability to detect complex samples, and achieves simple experimental operation and flexible design. , good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The present invention will be described in detail below in conjunction with the accompanying drawings and specific implementation examples.
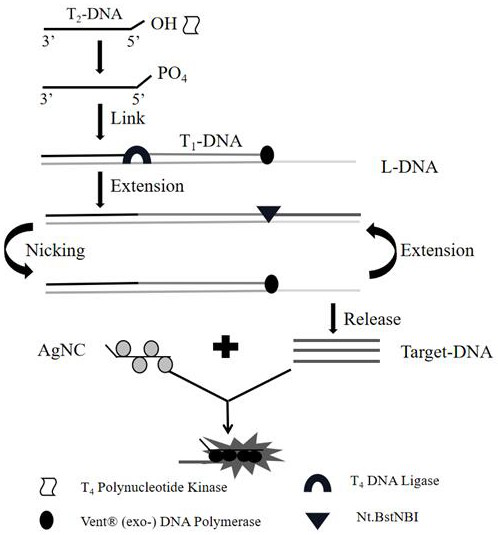
[0045] An oligonucleotide sequence-stabilized silver nanocluster (ssDNA / AgNCs) for high-salt, high-protein complex biological matrix samples T 4 polynucleotide kinase (T 4 PNK) activity detection method, characterized in that: after 1 mMZn 2+ , Ce 4+ , Pb 2+ Simple processing, high-salt, high-protein complex biological matrix samples can be directly used for sample detection without multiple filtration and dilution operations; ssDNA / AgNCs can still produce stable fluorescence output in high-salt, high-protein complex biological matrix samples, so that ssDNA / AgNCs has strong anti-interference ability and wider application range. The specific operation is as follows:
[0046] (1) with single chain T 2 -DNA as the starting point; first, T 4 In the presence of PNK, catalyzes the transfer and exchange of the phosphate group (Pi...
Embodiment 2
[0050] Quantitative detection of T in Hela cell extracts 4 PNK activity
[0051] 1. The characteristics of the preparation method of AgNCs rich in C sequence ssDNA stability include the following steps:
[0052] (a) Prepare 0.02 mM 100 mL pH7.0 PBS solution: weigh 0.7 g Na 2 HPO 4 and 1.7 g NaNO 3 , dissolved in deionized water, and diluted to 100 mL. Weigh 0.3 g NaH 2 PO 4 2H 2 O and 1.7 g NaNO 3 , dissolved in deionized water, and diluted to 100 mL. The first two solutions were mixed in any proportion to obtain a PBS solution with a final pH of 7.0;
[0053] (b) Prepare 0.3 mM 4 mL AgNO 3 Solution: weigh 40 mg AgNO 3 , dissolved in 8 mL deionized water, diluted 100 times to obtain 0.3 mM 4 mL AgNO 3 solution;
[0054] (c) Prepare 0.6 M 4 mL NaBH 4 Solution: weigh 18 mg NaBH 4 , dissolved in 8 mL deionized water, diluted 100 times to obtain 0.6 mM 4 mL NaBH 4 solution;
[0055] (d) 75 µL 0.02 mM PBS, 10 µL 50 µM ssDNA, and 10 µL 0.3 mM fresh AgNO 3 solution...
Embodiment 3
[0062] Quantitative detection of T in human serum 4 PNK activity
[0063] 1. The characteristics of the preparation method of AgNCs rich in C sequence ssDNA stability include the following steps:
[0064] (a) Prepare 0.02 mM 100 mL pH7.0 PBS solution: weigh 0.7 g Na 2 HPO 4 and 1.7 g NaNO 3 , dissolved in deionized water, and diluted to 100 mL. Weigh 0.3 g NaH 2 PO 4 2H 2 O and 1.7 g NaNO 3 , dissolved in deionized water, and diluted to 100 mL. The first two solutions were mixed in any proportion to obtain a PBS solution with a final pH of 7.0;
[0065] (b) Prepare 0.3 mM 4 mL AgNO 3 Solution: weigh 40 mg AgNO 3 , dissolved in 8 mL deionized water, diluted 100 times to obtain 0.3 mM 4 mL AgNO 3 solution;
[0066] (c) Prepare 0.6 M 4 mL NaBH 4 Solution: weigh 18 mg NaBH 4 , dissolved in 8 mL deionized water, diluted 100 times to obtain 0.6 mM 4 mL NaBH 4 solution;
[0067] (d) 75 µL 0.02 mM PBS, 10 µL 50 µM ssDNA, and 10 µL 0.3 mM fresh AgNO 3 solution, vorte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com