Application of HDS in predicting prognosis of gastric cancer patient, guiding postoperative adjuvant chemotherapy and predicting curative effect of immunotherapy
A technology for gastric cancer and patients, applied in the field of biomedicine, can solve problems such as difficult to precisely control changes in TME
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] Example 1 Calculation of histone deacetylase score (histone deacetylase score, HDS)
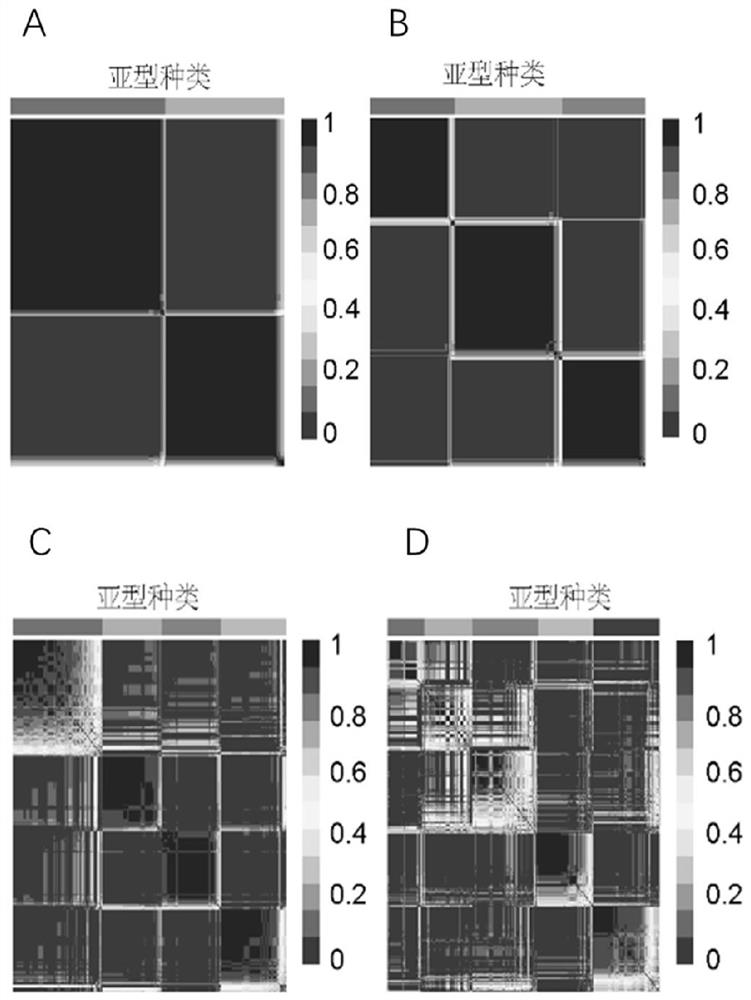
[0105] NMF cluster analysis was performed on the HDAC expression profiles of the ACRG cohort (also known as the GSE62254 cohort, which contains a large amount of complete clinical information), one of the largest datasets in GEO. It can be divided into three stable isoforms (HDAC clusters A, B and C) ( figure 1 ). The Limma software package was used to identify significantly differentially expressed genes between different HDAC clusters, P 1, and the high expression of this type of gene indicated poor prognosis, and this type of gene was defined as type B gene. Based on the expression of the above 103 genes, an HDS was constructed using principal component analysis (PCA) to quantify the score of each patient, and the first principal component (principal component 1) was used as the feature score. Use a method, such as GGI to define:
[0106] HDS=
[0107] PC1i represents the fea...
Embodiment 2
[0113] Example 2 Correlation analysis between HDS and prognosis of gastric cancer
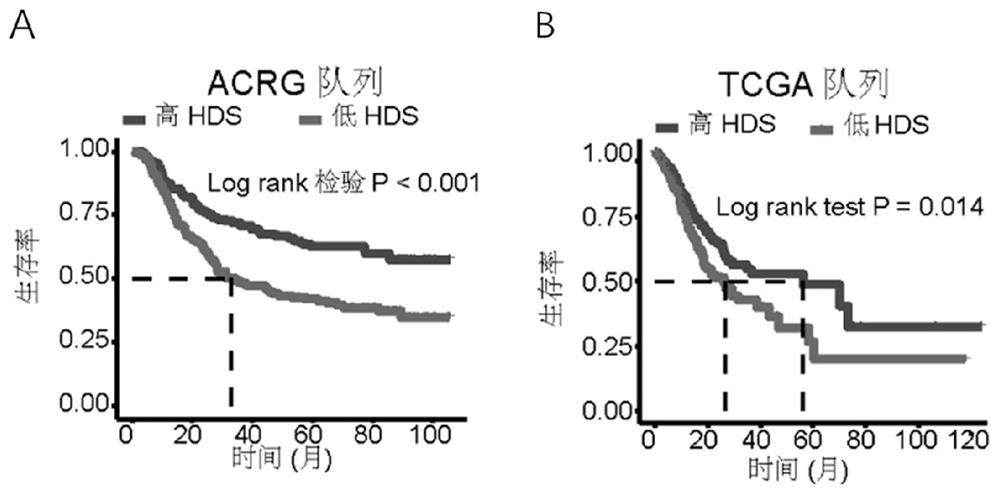
[0114] Gastric cancer samples in the ACRG cohort were divided into high HDS group and low HDS group according to the median, and the median was 0.08. The results showed that the high HDS group had a longer survival time than the low HDS group ( figure 2 in A).
[0115] Gastric cancer samples in the TCGA cohort were divided into high HDS group and low HDS group according to the median, with a median of 0.78. The results showed that the prognosis of low HDS was worse than that of high HDS ( figure 2 in B).
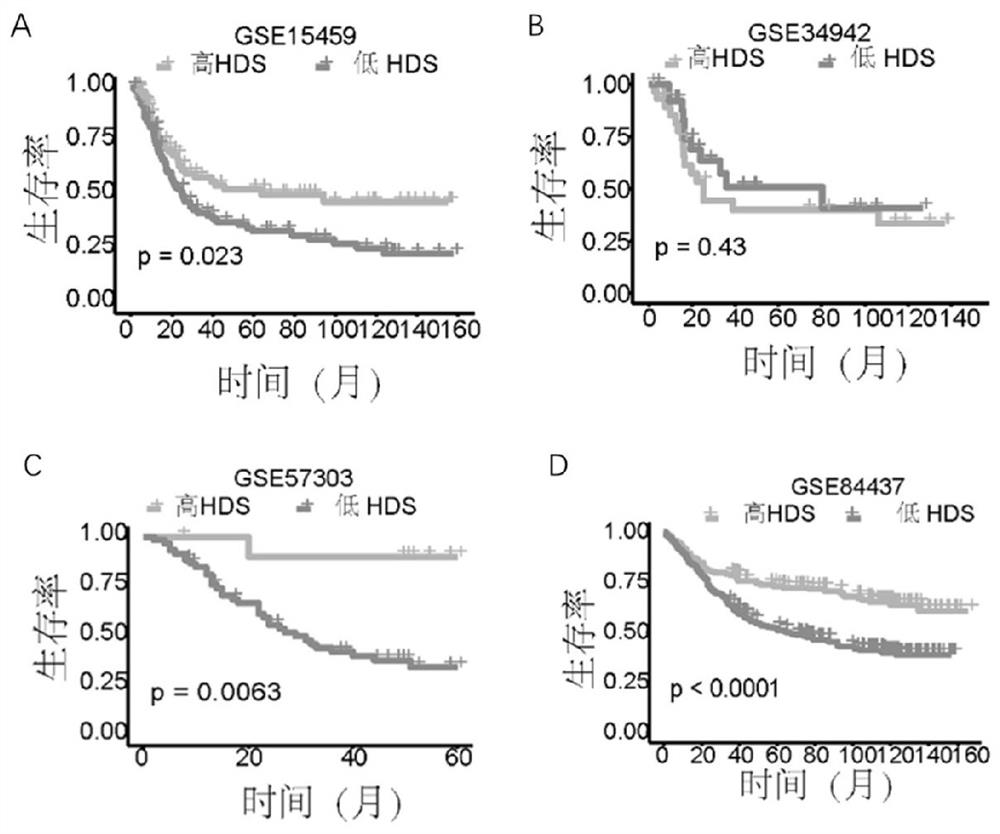
[0116] Analyzing the prognosis of high HDS and low HDS patients in different GEO cohorts, gastric cancer samples were divided into high HDS group and low HDS group according to the median, the median of GSE1545 was 0.65, the median of GSE34942 was 0.75, and the median of GSE57303 was 0.69GSE84437 The median is 0.106 ( image 3 ).
[0117] The above results indicated that HDS is reliable...
Embodiment 3
[0118] Example 3 Correlation analysis between HDS and the effect of immunotherapy in patients with gastric cancer
[0119] To elucidate the relationship between HDS and TME, the expression of HDS in six gastric cancer cohorts (TCGA-STAD, GSE15459, GSE34942, GSE57303, GSE62254, and GSE84437) was analyzed in relation to antigen presentation-related molecules, co-stimulatory molecules, co-inhibitory molecules, and angiogenesis Molecular correlations. It was found that HDS was positively correlated with the expression of most antigen-presenting molecules ( Figure 4 ), suggesting that the antigen presentation ability of patients with high HDS is significantly higher than that of patients with low HDS, and their ability to recognize and kill tumor cells in vivo is stronger. Further analysis found that HDS was positively correlated with the expression of co-stimulatory and co-inhibitory molecules ( Figure 4 ), suggesting that patients with high HDS have higher immunogenicity, but...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com