Novel method for detecting and quantifying mononucleotide variation
A single nucleotide variation and nucleotide technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of limited specificity of ARMS primers, large design limitations, and low mutation ratio. Achieve the effects of excellent reaction program compatibility, improved discrimination ability, and low mutation content
Pending Publication Date: 2022-03-29
XIAMEN ZEESAN BIOTECH
View PDF7 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0005] At present, ARMS technology has the following disadvantages: 1. ARMS technology is limited by the sequence at the end of the primer, and the design is relatively limited; 2. ARMS primers need to strictly control the reaction solution ratio and reaction temperature to meet the required detection requirements, and the reproducibility is low. Poor; 3. The specificity of ARMS primers is limited. Specificity refers to the concentration of non-detectable nucleotide sites that can be tolerated. The higher the concentration that can be tolerated, the lower the proportion of mutations that can be detected, which is more conducive to low mutations Accurate detection of content samples
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
[0028] In order to make the technical problems, technical solutions and beneficial effects to be solved by the present invention clearer and clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments.
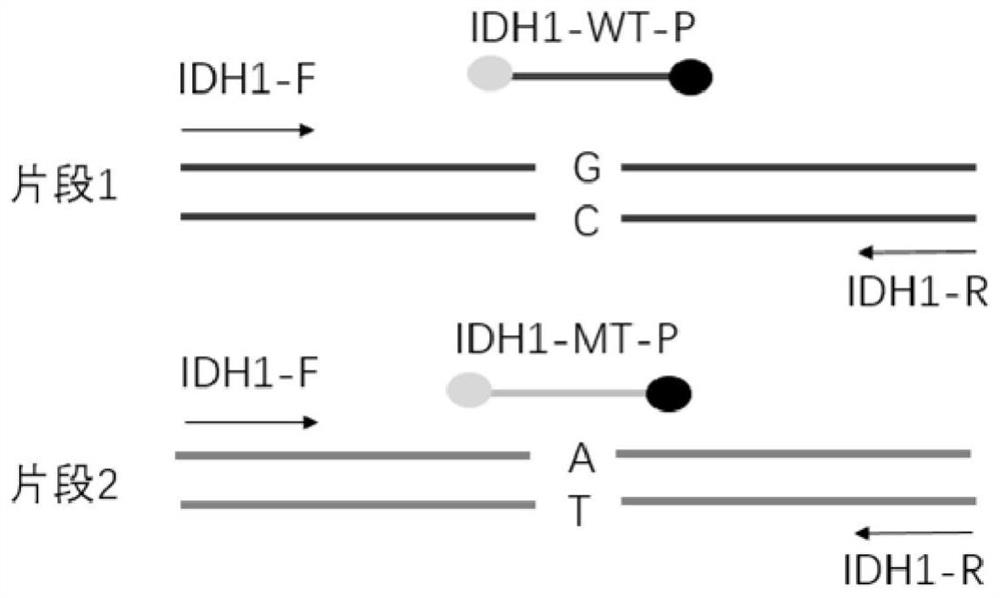
[0029] 1. Design of primers and probes
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Login to View More
Abstract
A novel method for detecting and quantifying mononucleotide variation is characterized in that a probe is combined with locked nucleic acid (LNA) modification to improve the difference between Tm1 and Tm2, the Tm1 is the Tm value when the probe is combined with a completely matched sequence, the Tm2 is the Tm value when the probe is matched with a mononucleotide variation sequence, and the modified probe is used for distinguishing and quantifying mononucleotide variation sites. The nucleotide in the probe is subjected to LNA locked nucleic acid modification, and the temperature difference between Tm1 and Tm2 is improved, so that the distinguishing capability of the probe on a single nucleotide variation sequence is improved, and a template with lower mutation content can be detected.
Description
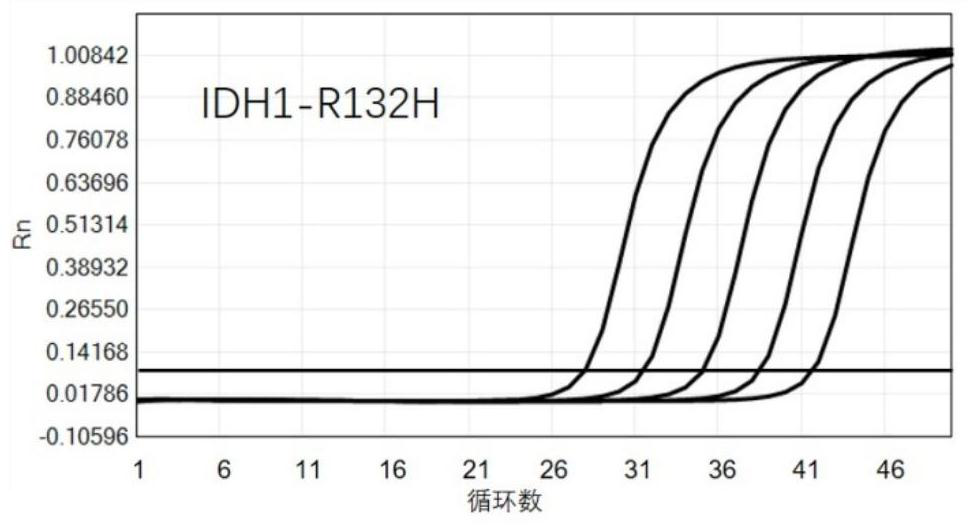
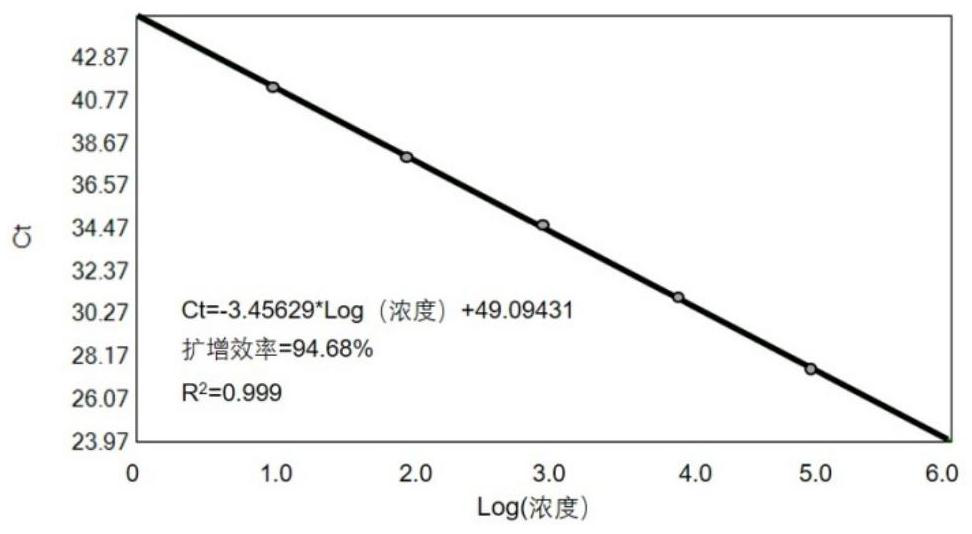
technical field [0001] The invention relates to the field of biotechnology, in particular to a new method for detecting and quantifying single nucleotide variation. Background technique [0002] The identification of single nucleotide variation is of great significance in drug monitoring and other aspects. For example, a specific gene mutation is an important biomarker for tumor-targeted therapy, and gene mutation detection can detect drug-resistant mutations as early as possible before the disease progresses, helping doctors make clinical treatment decisions and strive for treatment opportunities for patients. [0003] At present, the detection of single nucleotide variation mainly uses a real-time fluorescence quantitative PCR (polymerase chain reaction, polymerase chain reaction) platform. The real-time fluorescent quantitative PCR platform is a method of measuring the total amount of products after each cycle of polymerase chain reaction (PCR) with fluorescent chemicals...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/6851C12Q1/6858
CPCC12Q1/6851C12Q1/6858C12Q2531/113C12Q2563/107C12Q2545/114C12Q2525/10
Inventor 倪润芳郭婷婷赵菁康金妹李官林
Owner XIAMEN ZEESAN BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com