Probe combination for diagnosing fibula amyotrophy, kit and application
A technology of Charcot-Marie-Tooth disease and a kit, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve problems such as missed diagnosis, improve accuracy, improve accurate diagnosis rate, reduce time and The effect of financial burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
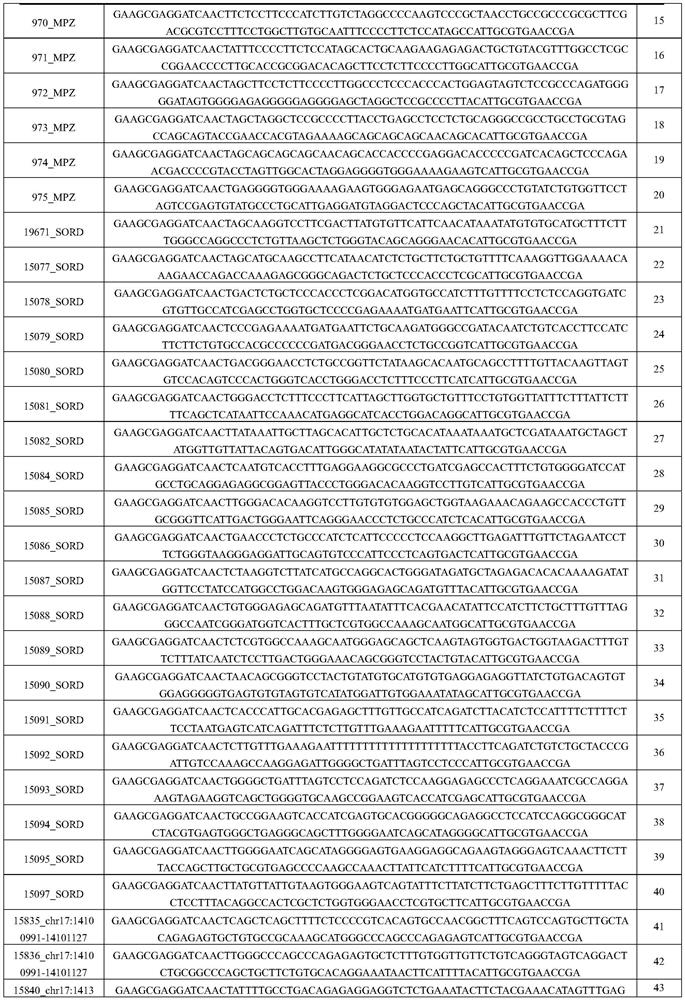
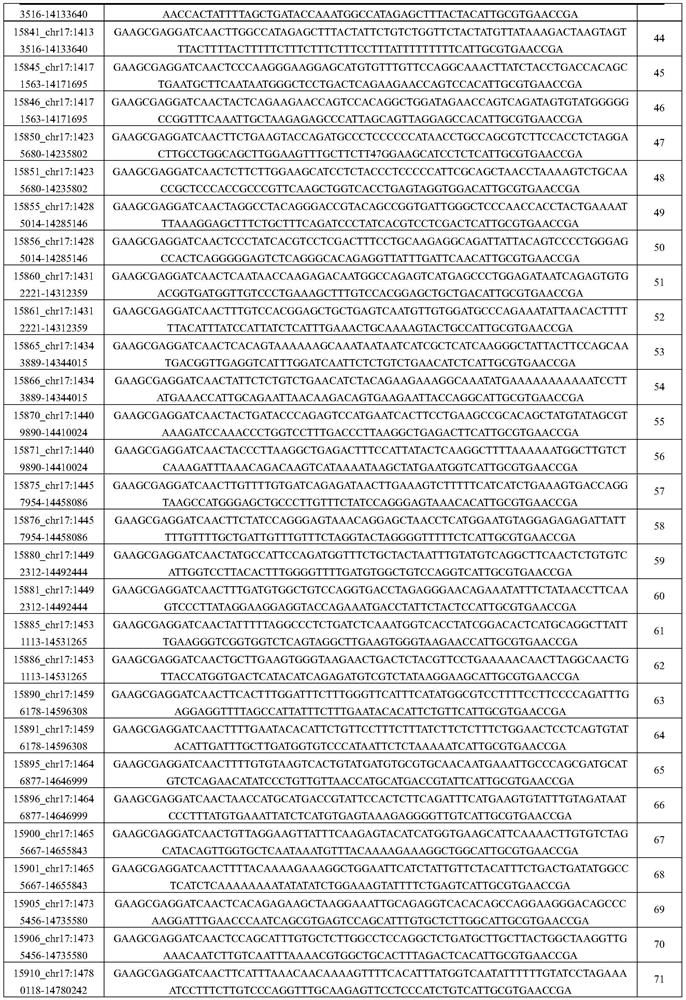
[0049] Embodiment 1 probe design
[0050] This embodiment uses the following methods to design specific probes for CMT-related gene detection:
[0051] 1) The probe covers the exon region and 50bp at both ends of the exon;
[0052] 2) For regulatory regions other than exons, use 100 bp as a sliding window. When there are 2 or more known pathogenic mutations in one window, keep this interval and design related probes;
[0053] 3) The probe design uses the shingled style, and each base in the reference interval is covered by at least 3 probes;
[0054] 4) For the target gene with CNV, add a 200bp CNV-plus probe interval at every interval of 1-3k. This probe interval needs to include the SNP positions with a mutation frequency of 20%-80% in the East Asian population in the gnomad population database point and there is no similar sequence on the genome in this interval (using 200bp as the sliding window, the whole genome comparison is performed, and there is no sequence with a s...
Embodiment 2
[0076] Example 2 Target Region Capture Sequencing
[0077] In this example, the capture probe set provided in Example 1 is used, and the sequence capture technology based on liquid phase hybridization is used to enrich the target sequence. The technical route of liquid phase hybridization is: (1) prepare a hybridization probe library, (2) use the probe to enrich the target gene, (3) sequence the enriched DNA sequence with a high-throughput sequencer.
[0078] The use of liquid phase hybridization technology can effectively enrich the target gene, reduce the requirement of sample size, and better control the cost of sequencing. The library construction method comprises the following steps:
[0079] 1. DNA extraction and fragmentation
[0080] DNA was extracted from 4 peripheral blood samples using the QIAamp DNA Blood Mini Kit.
[0081] Use the Bioruptor Pico DNA interrupter, after the temperature of the cold cycler drops to 4°C, set the parameters ON for 30s, OFF for 30s as...
Embodiment 3
[0161] Example 3 Sample Detection
[0162]20 samples were selected for detection by the method shown in Example 2, and WES and MLPA were used to conduct parallel comparison experiments on the CNV of the PMP22 gene of the samples. The kit used for WES is: SureSelect human whole exon V7 exome; the kit used for MLPA is: MRC-Holland P405 detection kit.
[0163] The results are shown in Table 11, which shows that the detection result of the method in Example 2 has a coincidence rate of 100% with MLPA, which is higher than the result based solely on the Reads depth analysis.
[0164] Table 11 Sample test results
[0165]
[0166]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com