Molecular marker for quantifying lactobacillus acidophilus and method for absolutely quantifying bacterial community composition in yellow wine fermentation process
A technology of molecular markers and absolute quantification, applied in the field of bioengineering, can solve the problems of determining the complexity of the original internal standard, the complex microbiome of rice wine, and the inability to use it, and achieve the effect of convenient operation, wide application range, and overcoming the inconvenience of operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0012] Example 1. Design of Lactobacillus acetotolerans-specific primers
[0013] 1. Synthetic primers: based on the Lactobacillus acetotolerans (Lactobacillus acid-tolerant Lactobacillus) genome as the basic data, a specific nucleic acid sequence was screened, and then a specific nucleic acid sequence was found on the genome of Lactobacillus acetotolerans through manual search and comparison, such as SEQ ID NO: 3 Shown), this sequence has only one copy on the Lactobacillusacetotolerans genome, and based on this sequence, primers LA-F (5'-GTGCGTTACGGAGGAGTA-3', SEQ ID NO: 1) were designed on the basis of this sequence; LA-R (5' -CGATCATTGAATCGTCAGG-3', SEQ ID NO: 2), the structure of the primer itself meets the requirements.
[0014] 2. Specificity verification:
[0015] (1) Choose to buy microorganisms in 18 kinds of rice wine for primer specificity test [Lactobacillusplantarum (CGMCC 1.568); Lactobacillus brevis (CGMCC 1.214); Lactobacillus acetotoolerans (ATCC 27745); Lact...
Embodiment 2
[0020] Embodiment 2. A kind of test kit of quantitative acid-resistant Lactobacillus
[0021] The components are: LA-F (5'-GTGCGTTACGGAGGAGTA-3', SEQ ID NO:1), LA-R (5'-CGATCATTGAATCGTCAGG-3', SEQ ID NO:2), AceQ Universal SYBR Green qPCR MasterMix, standard DNA and ddH 2 O. The standard product DNA is a plasmid containing the molecular marker shown in SEQ ID NO:3.
Embodiment 3
[0022] Example 3. Quantitative method of internal standard Lactobacillus acetotolerans
[0023] Using the Lactobacillus acetotolerans genome as a template, use specific primers LA-F, LA-R to amplify the target fragment to obtain the product of SEQ ID NO: 3. The amplified product is cloned into pMD19-T and transformed into Escherichia coliJM109. Colony PCR verifies the transformed Positive clones; use a plasmid extraction kit to extract the plasmid, and quantify the concentration of the plasmid with a micro-ultraviolet spectrophotometer;
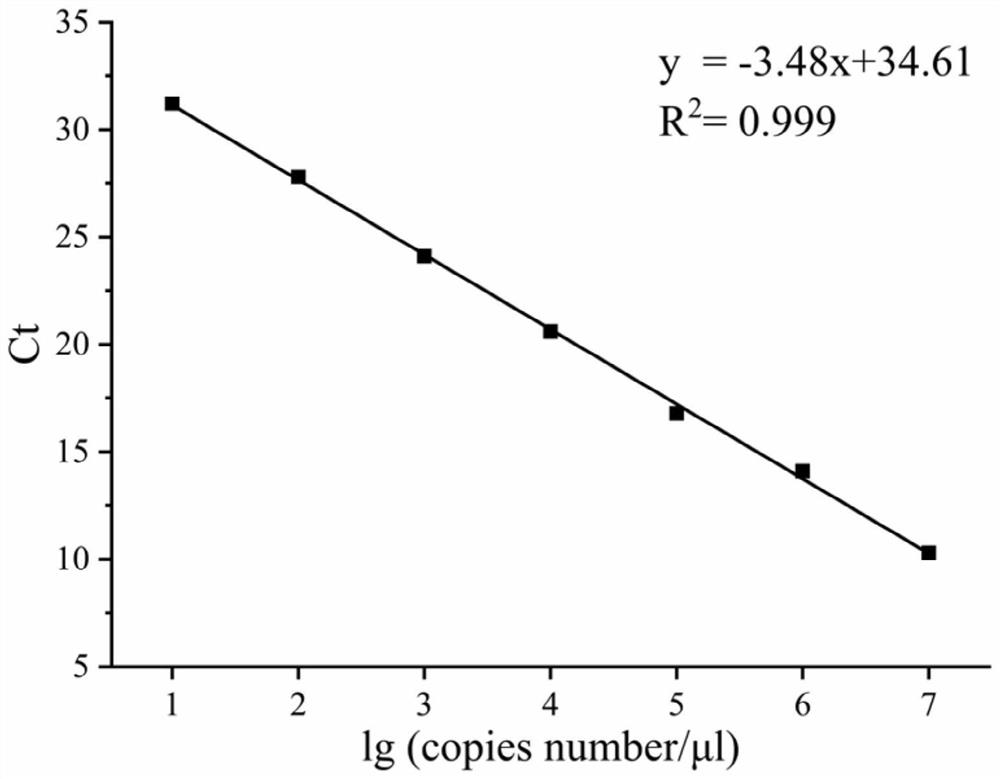
[0024] The quantified plasmids were diluted tenfold, and 8 gradients were taken respectively, and the 8 gradients were used for qPCR. PCR system: 1 μL of template, 0.4 μL of upstream and downstream primers, 10 μL of AceQ Universal SYBR Green qPCR Master Mix, supplemented with double distilled water to The final volume is 20 μL; PCR conditions: 98°C for 1 min, 98°C for 10 s, 61°C for 1 min, 40 cycles.
[0025] standard curve as image 3 As s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com