Specific selective amplification and multiplex PCR method and application
A specific, multiple technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
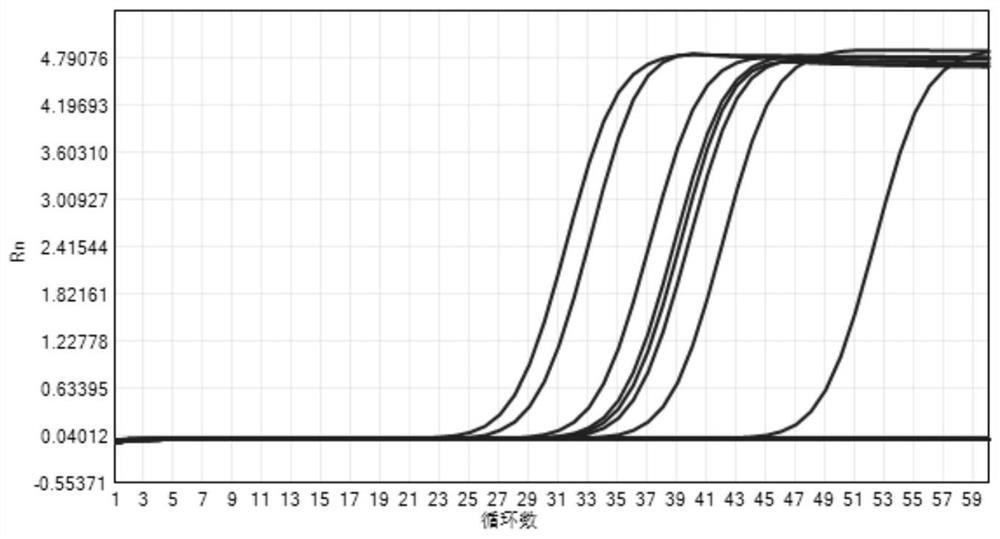
[0029] Use the PCR dye method to compare the non-specific amplification and specific amplification of the present invention in conventional PCR
[0030] Conventional primer pairs RNaseP1(seq1), RNaseP2(seq2) and modified primer pairs RNaseP1M(seq3), RNaseP2M(seq4) were synthesized, and tested by conventional qPCR system and qPCR system with endonuclease IV, respectively.
[0031] The specific PCR reaction system is:
[0032] a) Conventional group: 10mM Tris-HCL (pH8.7), 50mM KCL, 1×EvaGreen (Biotium), 40U / mL Hotstart Taq DNA polymerase (Wethink), 2U / mL UDG (Roche), 0.2mM dATP, 0.2mM dCTP, 0.2 mM dGTP, 0.4 mM dUTP, 0.2 μM RNaseP1, 0.2 μM RNaseP2.
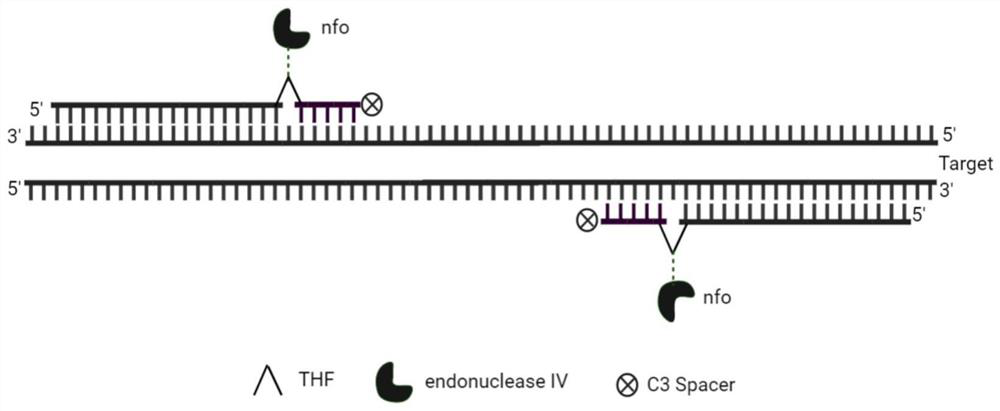
[0033] b) Modification group: 10mM Tris-HCL (pH8.7), 50mM KCL, 1×EvaGreen (Biotium), 40U / mL Hotstart Taq DNA polymerase (Wethink), 2U / mL UDG (Roche), 0.2mM dATP, 0.2mM dCTP, 0.2mMdGTP, 0.4mM dUTP, 0.2μM RNaseP1M, 0.2μM RNaseP2M, 20U / mL Tth nfo enzyme (NEB)
[0034] The primer sequences are as follows:
[0035] Seq1(RNaseP1): AGA ...
Embodiment 2
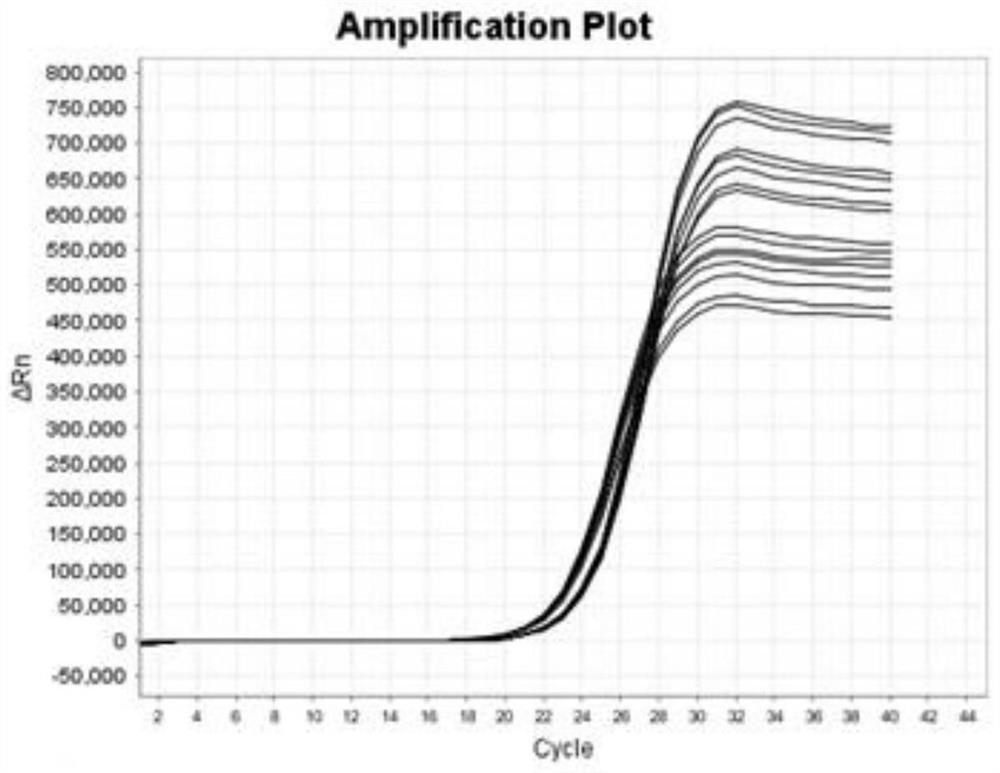
[0045] Blood Screening HBV / HCV / HIV-1 / HIV-2 / internal standard (IC) 5-color fluorescent 8-fold RT-PCR system
[0046] Specifically:
[0047] Two pairs of primers and two FAM probes for HBV, targeting S gene and C gene respectively, one pair of primers for HCV and one CY5 probe, two pairs of primers for HIV-1 and two ROX probes, targeting GAG gene and POL gene respectively, HIV-2 Two pairs of primers and two VIC probes are aimed at the GAG gene and LTR region respectively. The internal standard adopts exogenous pseudovirus, and one pair of primers and one TAMRA probe. A total of 8 pairs of primers (merged bases in the primers) and 8 TaqMan probes (merged bases in the probes), a total of 24 oligonucleotides, the probes are modified at both ends, cannot be extended, and only have specificity Hybridization binding, after being cleaved by a thermostable polymerase, it causes a fluorescence change function. The probe remains unchanged, and the 8 pairs of primers are respectively u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com