Kit for detecting mutations causing genetic disorders
A technology for kits and diseases, applied in the field of kits for detecting mutations that cause genetic diseases, can solve problems such as time-consuming and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
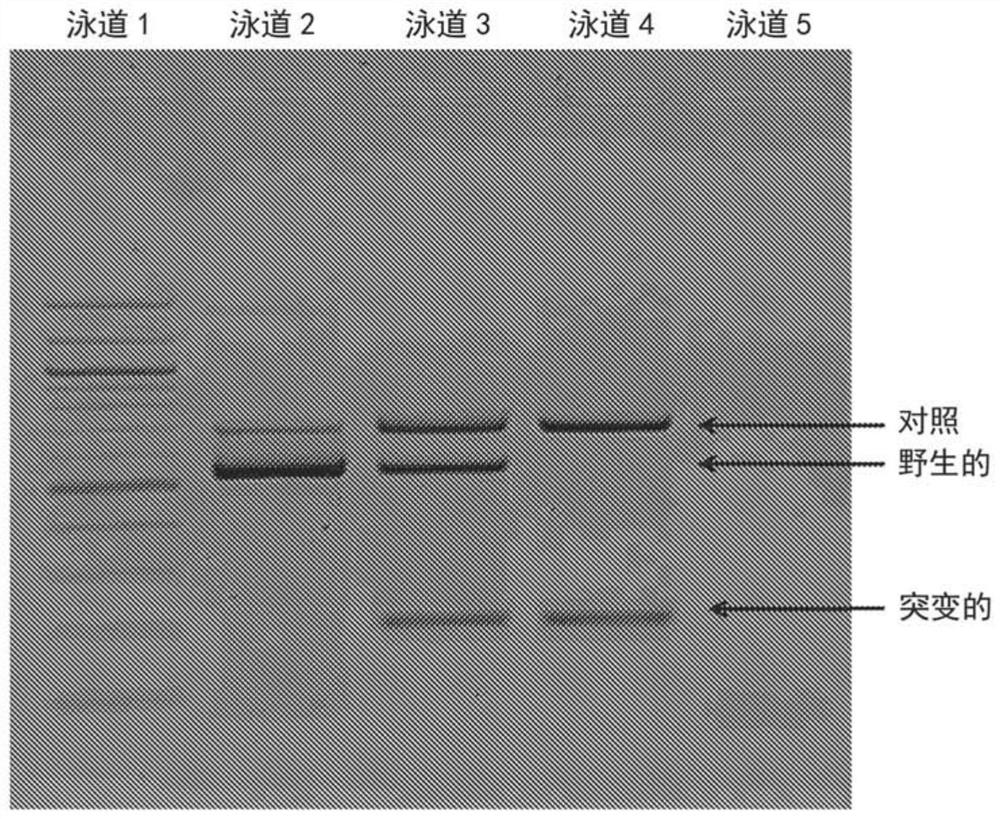
[0061] The ARMS-PCR assay detects sickle cell anemia-causing mutations using unprocessed human dried blood spots (DBS) in a single reaction.
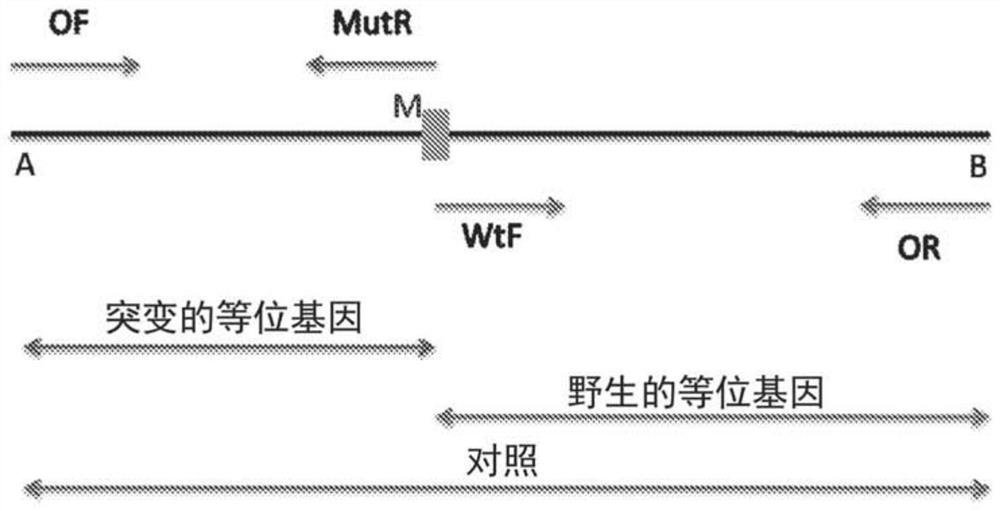
[0062] To test the capabilities of the method developed in the present invention, untreated human dried blood spots were used. Multiple primers were designed to detect wild-type genotype (A / A), mutant genotype (T / T) and heterozygous genotype (A / T) of Cd6 T>A mutation in HBB gene. External forward primer [50nM, 20nt, 5'd (ACC TCA CCC TGT GGA GCC AC) 3'] (SEQ ID NO: 1) and external reverse primer [50nM, 20nt, 5'd (TCA TTC GTC TGT TTC CCA TT)3'] (SEQ ID NO:2), allele-specific primer, internal forward of A allele [50nM, 20nt, 5'd(ATG GTG CAT CTG ACT CCT GA)3']( SEQ ID NO:3), and the internal reverse of the T allele [50nM, 21nt, 5'd(CAG TAA CGG CAG ACT TCT CCA)3'] (SEQ ID NO:4) for detection of specific alleles . Templates for all three genotypes (mutant, heterozygous and wild type) were used. The reaction mixture contained 1X buffer, in...
Embodiment 2
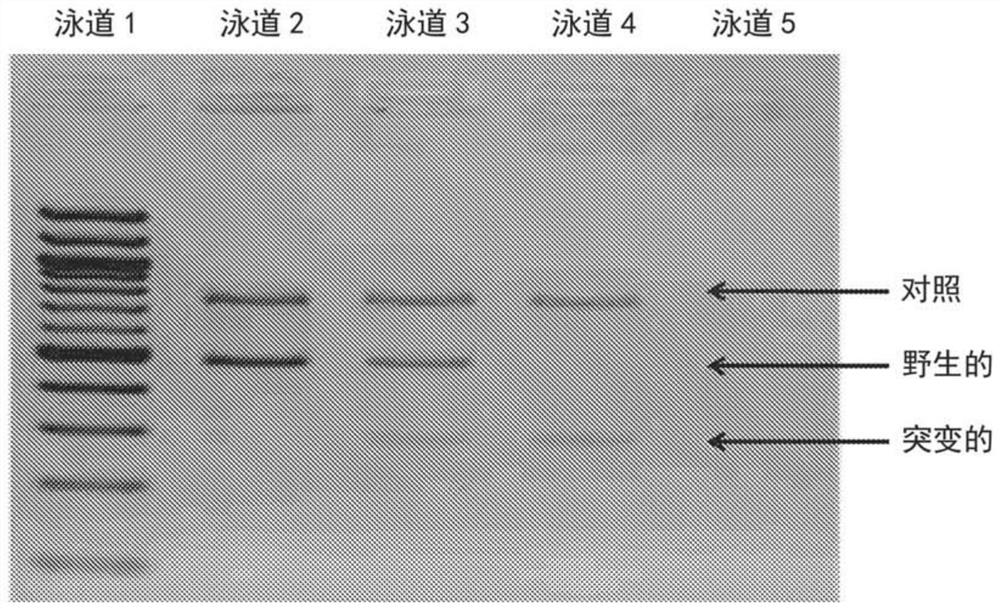
[0065] The ARMS-PCR assay was used to detect 5 common mutations causing β-thalassemia using untreated human dried blood spots separately under a single PCR condition.
[0066] Such as Figure 4 As shown, an ARMS-PCR assay for β-thalassemia mutations was established using untreated human dried blood spots under one common PCR condition. Allele-specific primers have been used so that both alleles can be detected in the same reaction. A number of primers were designed to detect wild-type, mutant and heterozygous genotypes. Used external forward primer 1 [50nM, 20nt, 5'd (ACC TCA CCC TGT GGA GCC AC) 3'] (SEQ ID NO: 1), external forward primer 2 [50nM, 26nt, 5'd (GTA CGG CTG TCA TCA CTT AGA CCT CA) 3'] (SEQ ID NO: 11) and external reverse primer [50nM, 20nt, 5'd(TCA TTC GTC TGT TTC CCA TT) 3'] (SEQ ID NO: 2 ). For the IVS1>5G>C mutation in the HBB gene, an allele-specific primer for G internal forward [50nM, 20nt, 5'd(TGA GGCCCT GGG CAG GTA GG)3'] (SEQ ID NO :6) and an interna...
Embodiment 3
[0069] Allele-specific PCR assay for the detection of spinal muscular atrophy (SMA)-causing exonic deletions using untreated human DBS in a single reaction.
[0070] To detect deletions in the SMN gene for the diagnosis of SMA using unprocessed human dried blood spots, multiple allele-specific primers were designed. This will detect deletions of exon 7 and exon 8 in the SMN gene and simultaneously differentiate between SMN1 and SMN2 copies of the SMN gene in one reaction. Forward primer [50nM, 22nt, 5'd (TTT ATT TTC CTTACA GGG TTT C) 3'] (SEQ ID NO: 22) of exon 7 was used, internal reverse primer [50nM, 24nt, 5'd (GTG AAA GTA TGTTTC TTC CAC GTA)3'] (SEQ ID NO:24), internal forward primer for exon 8 [50nM, 25nt, 5'd(CTG GCATAG AGC AGC ACT AAA TGA C)3'] (SEQ ID NO:27), exon 8 specific reverse primer of SMN1 [50nM, 19nt, 5'd((TGG CCT CCC ACC CCC AAC C)3'](SEQ ID NO:23). As internal As a control, a control forward primer [50nM, 22nt, 5'd(AAG GAC AAT GGG AAC ACT CTC T)3'] (SEQ ID...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com