Method for improving eukaryotic gene editing efficiency and application thereof
A technology of gene editing and eukaryotes, applied in biochemical equipment and methods, microorganisms, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
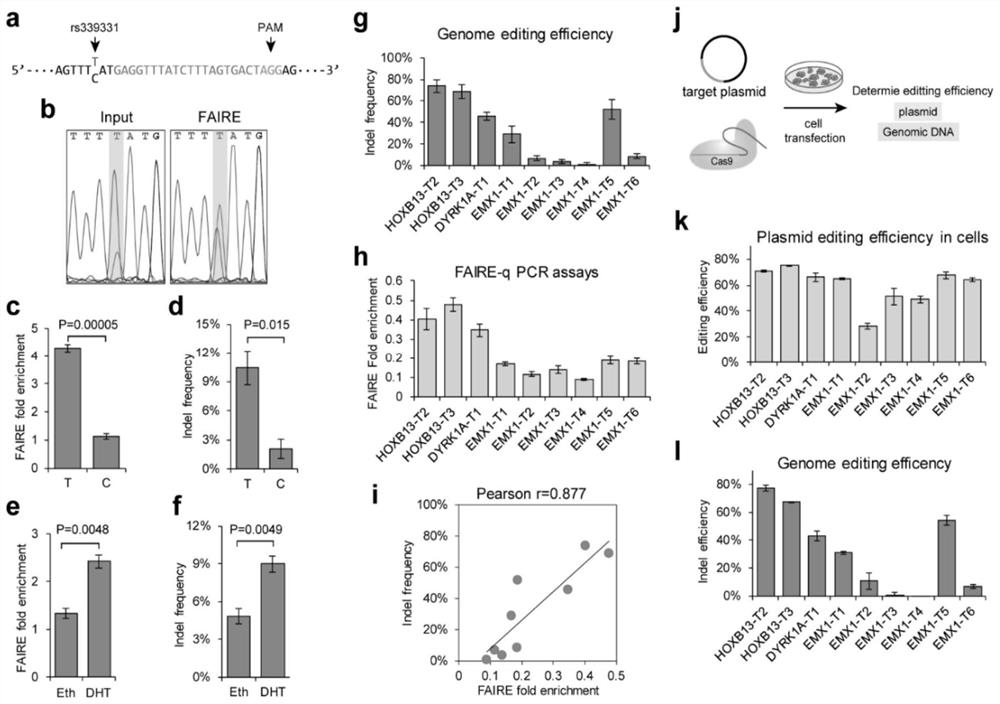
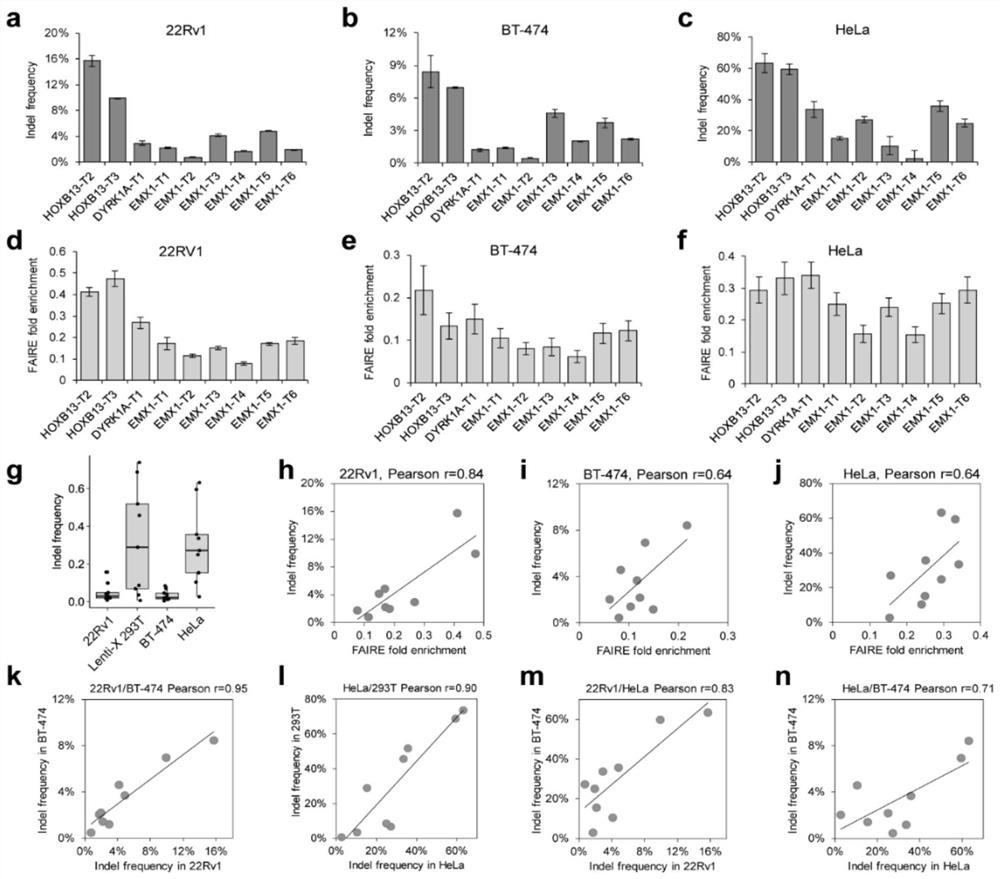
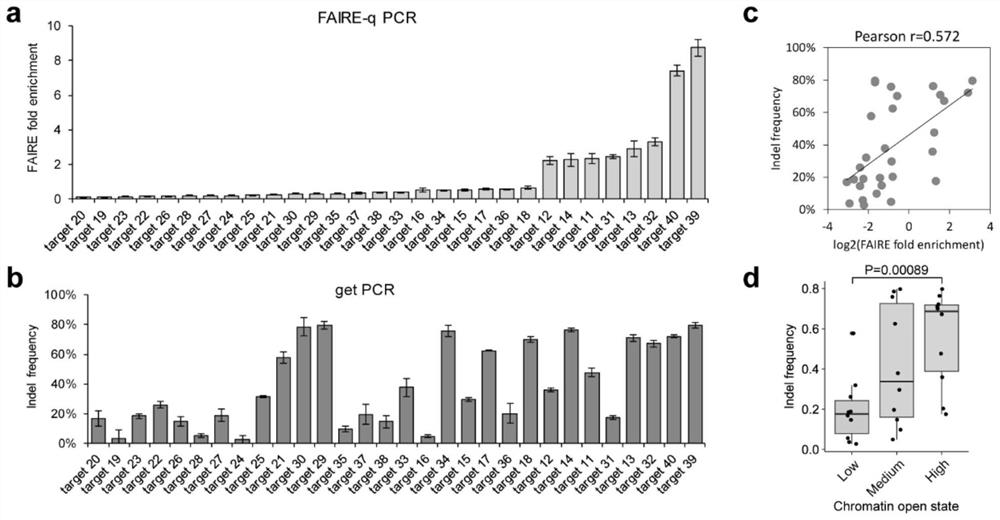
[0072] 1.1 Genome editing efficiency is related to chromatin opening state
[0073] The rs339331 locus is T / C heterozygous in two prostate cancer cell lines, VCaP and 22Rv1, and previous studies revealed that these two alleles have different chromatin opening states. Directly compare the genome editing efficiency of two different alleles near the rs339331 site ( figure 1 a), allowing the present invention to strip the potential impact of sgRNA target sequence differences when studying the correlation between genome editing efficiency and chromatin open state. Sanger sequencing results of FAIRE DNA in VCaP cells ( figure 1 b) Sanger sequencing results of FAIRE DNA in and 22Rv1 cells both showed T allele enrichment. FAIRE qPCR analysis of these two cell lines using allele-specific primers confirmed that the T allele has a more open chromatin state than the C allele ( figure 1 c). In VCaP ( figure 1 d) The indel frequency of the T allele is significantly higher than that of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com