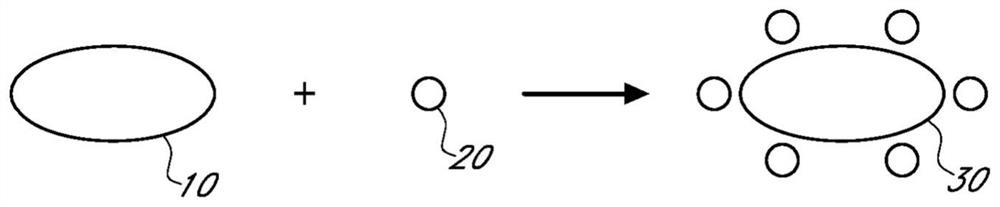
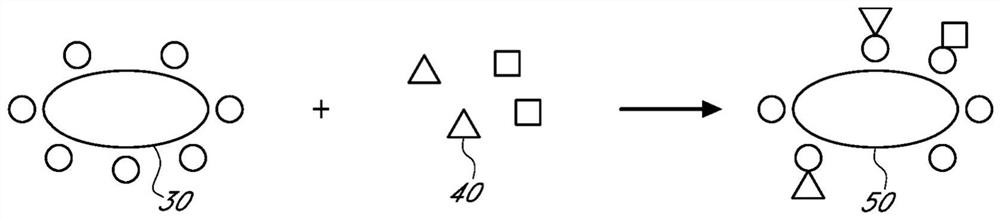
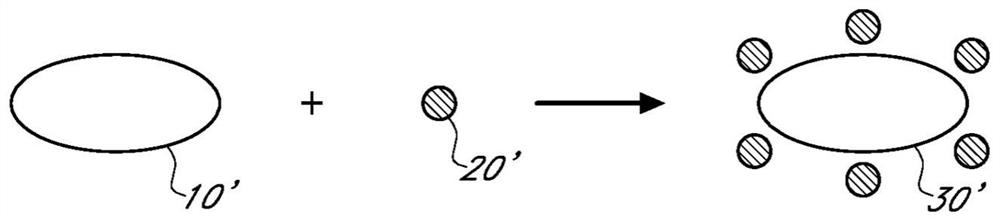
Sample preparation compositions, devices, systems, and methods
A composition and sample technology, which is applied in the preparation methods of peptides, chemical instruments and methods, separation methods, etc., can solve the problems of poor protein recovery, time-consuming and boring, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0168] Aspects of the present teachings can be further understood from the following examples, which should not be construed to limit the scope of the present teachings in any way.
example 1
[0169] Example 1: Preparation and Testing of Compositions
[0170] A composition for isolating or extracting one or more small molecules from a sample is prepared comprising at least one size exclusion support and at least one moiety that can be associated with one or more small molecules, and applying the The composition described above was tested.
[0171] In some embodiments, the reagents include small molecules that can associate with small molecules through charge interactions, hydrophobic interactions, or any other interactions to associate with these small molecules and thereby remove these small molecules from the sample, while allowing for exclusion and collection Exemplary size exclusion supports are modified by portions of functional groups of larger biomolecules in the sample.
[0172] In this example and the following examples, preparations were made for dyes (in the molecular weight range of about 700 Da-1100 Da), biotin and its derivatives (in the molecular wei...
example 1A
[0174] Example 1A: Preparation and Testing of Chemical 1
[0175] Chemical 1: Partial immobilization of size exclusion support with branched polyethyleneimine: Size exclusion for hydroxyethyl cellulose resins including 7K and 40K size exclusion ranges, e.g. Zeba 40K and Zeba 7K spin desalting columns from ThermoScientific, as described below Two examples of supports were modified. Periodate oxidation of vicinal diols positioned on these size exclusion support columns produces aldehyde groups. Polyethyleneimine and diethylenediamine with primary amines are reacted with the resulting aldehydes using Schiff base chemistry. Polyethyleneimine and diethylenediamine were prepared in PBS, the pH was adjusted to a range of 8.0-8.5, and the polyethyleneimine and diethylenediamine were reacted with an oxidized Zeba column. These compositions are hereafter referred to as Zeba 7K-PEI (polyethyleneimine) and Zeba40 K-PEI (polyethyleneimine).
[0176] The same chemical modification was ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com