System for determining sample type based on DNA methylation level, readable medium and application thereof
A technology of methylation and methylation rate, which is applied in the field of determining sample types based on DNA methylation level, can solve the problems of rare tumor DNA, difficulty in methylation rate, and too much loss of details, achieving reliable results and solving problems The effect of low detection rate and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
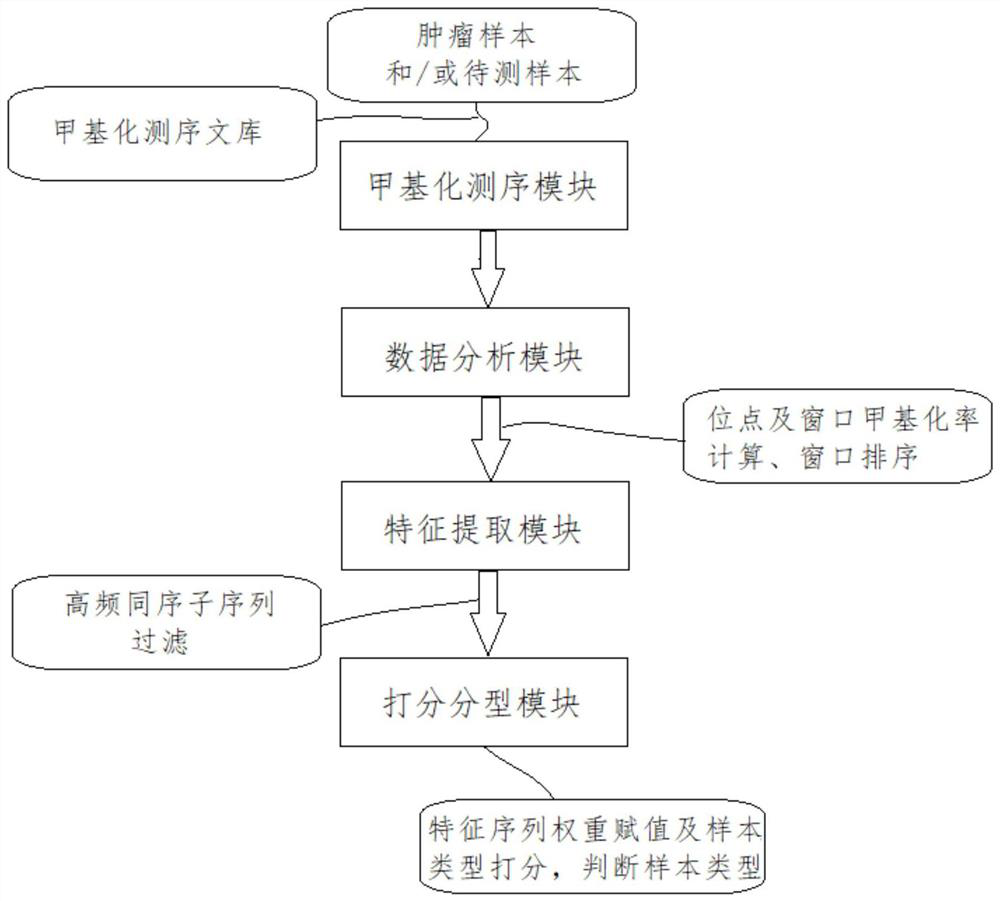
[0059] like Figure 1~3 As shown, this embodiment includes: a methylation sequencing module, a data analysis module, a feature extraction module and a scoring and typing module, wherein: suspected bladder cancer patients and confirmed bladder cancer patients (tumor types) that have not yet received treatment are collected and Peripheral fluid samples of healthy people (healthy type) were subjected to DNA methylation sequencing using the BGI-seq500 sequencing platform, and methylation sequencing libraries were constructed respectively.
[0060] The methylation sequencing module performs DNA methylation sequencing on the constructed methylation sequencing library to obtain sequencing data; the sequencing data includes several sequencing reads.
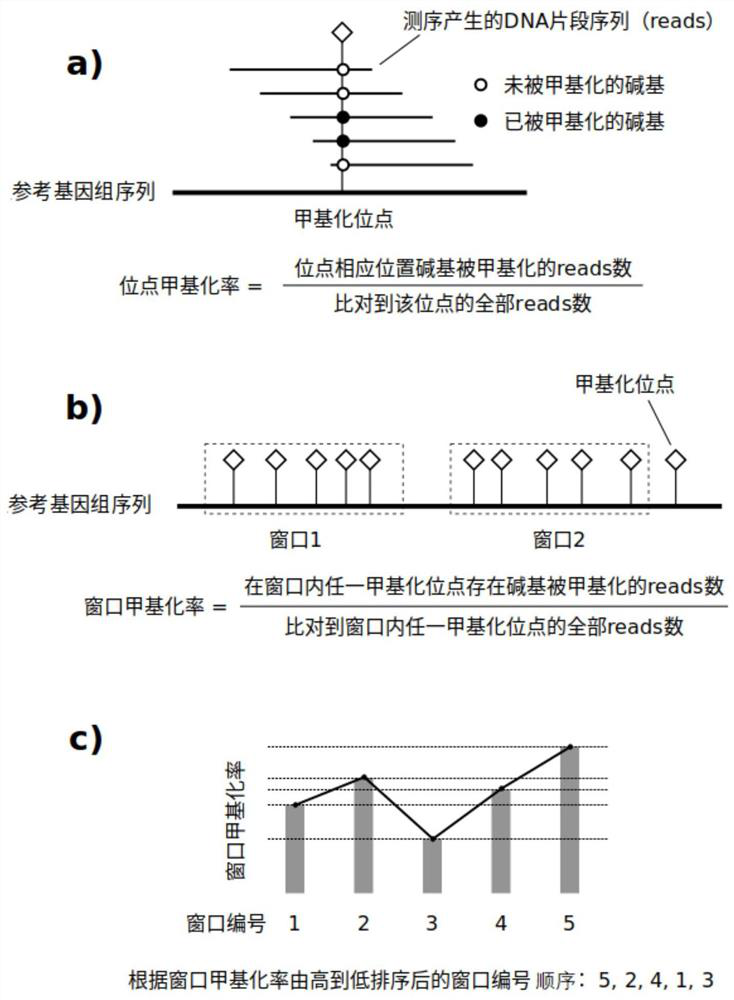
[0061] The data analysis module is used to divide the reference genome into windows, and calculate the window methylation rate of each sample based on the sequencing reads of the methylation sequencing library of the sample to be tested,...
Embodiment 3
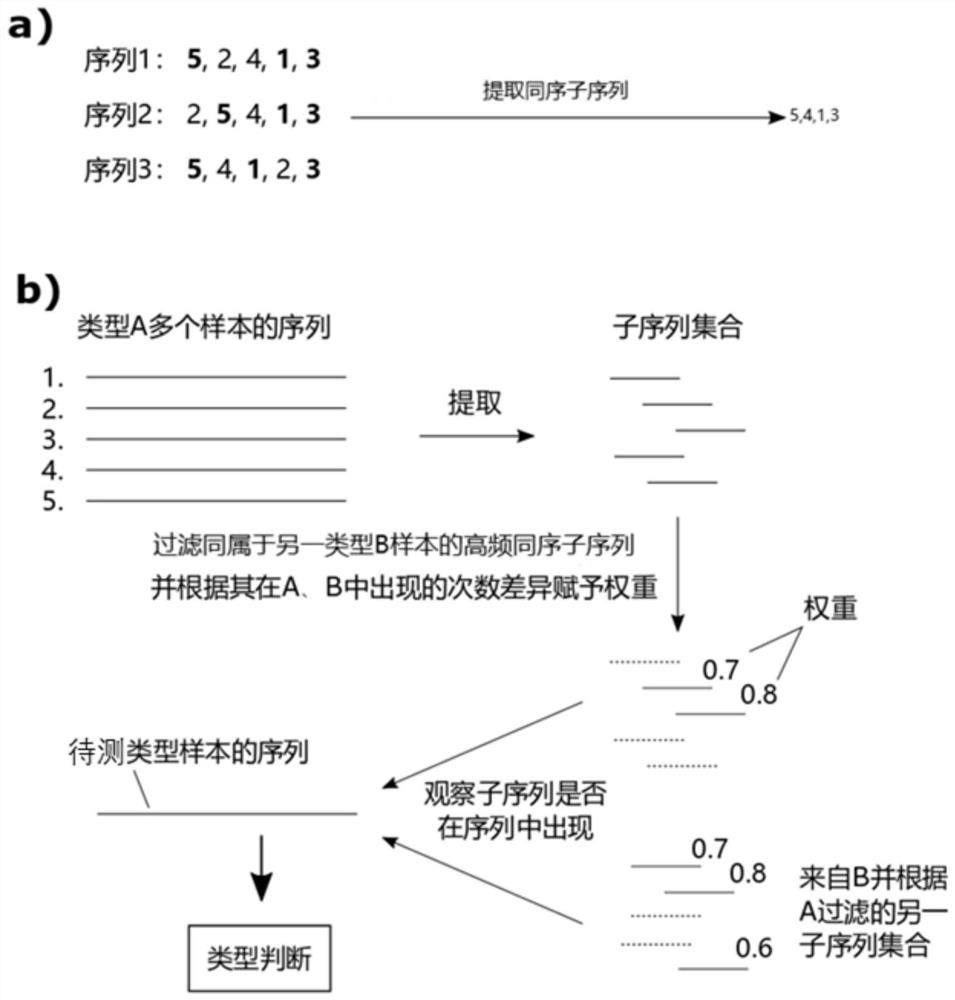
[0093] The source of samples in this example is the urine of donors (30 bladder cancer patients and 10 healthy individuals). Methylation sequencing used conventional methylation library construction, BGQ-seq500 sequencing platform, and sequencing depth 30×. The number of methylation sites included in each window is set to 50, the threshold of the proportion of high-frequency homosequence subsequences is 50%, and the weight is the reciprocal of the p value of Fisher's exact test (transformed using the log function and multiplied by -1) , and perform 10-fold cross-validation for a total of 10 rounds of testing.
[0094] In this example, 50 methylation sites as a window is the optimal parameter for this batch of data. Generally speaking, the more methylation sites a window contains, the more stable the statistical features are, because it refers to more sites, but the more details are lost, because the same window only considers the overall index (window A methylation rate), whi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com