Method for comparing expression amount of idential gene in different origin using base sequence determination method
A technology of base sequence and expression, which is applied in the field of quantitative comparison of the relative expression of the same gene in tissues or cells of different origins, and achieves the effects of easy instrumentation, convenient operation and low price.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] [Example 1] Determination of the difference in the expression of P53 gene in human normal tissue, brain cancer tissue and liver cancer tissue.
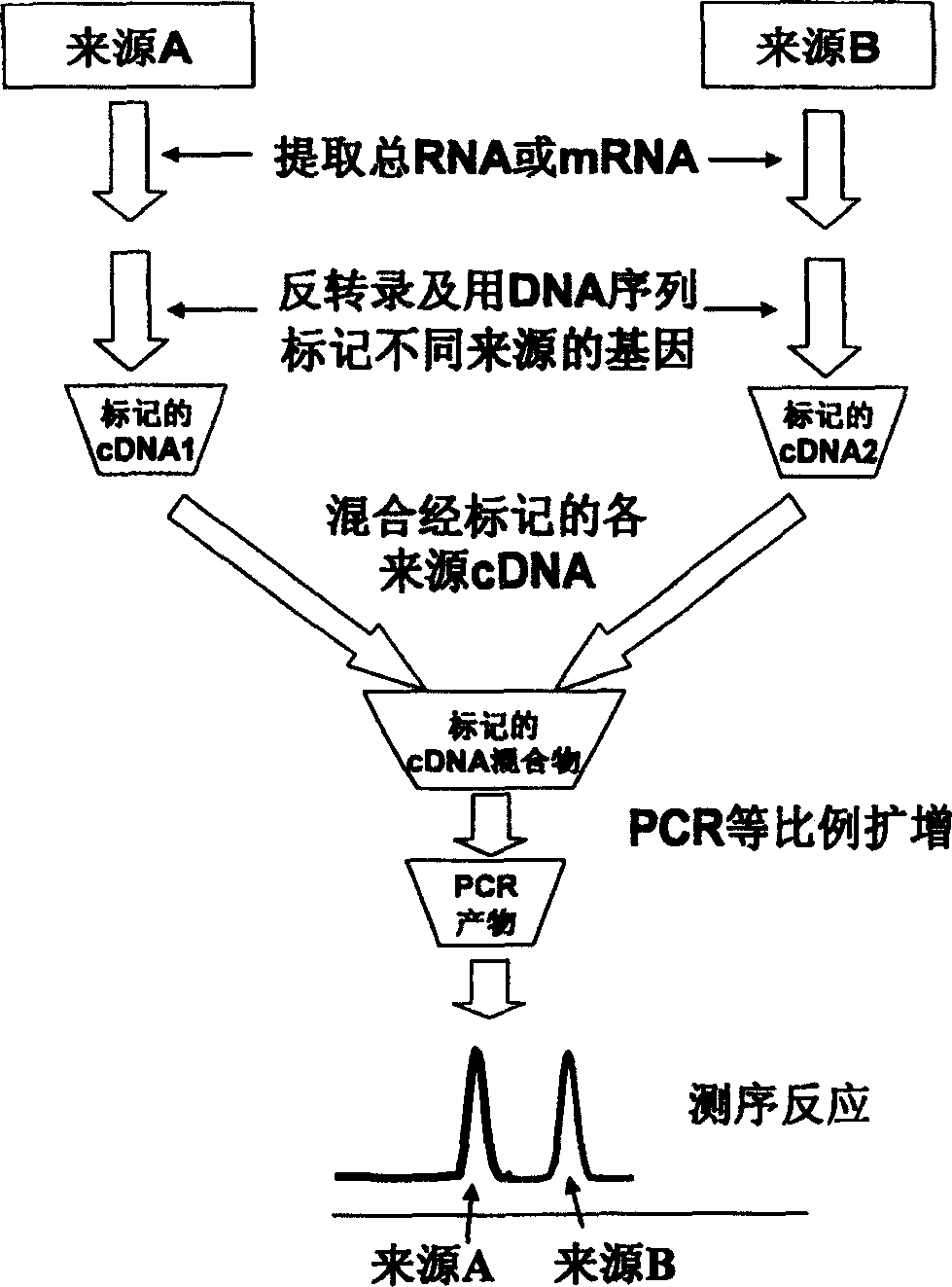
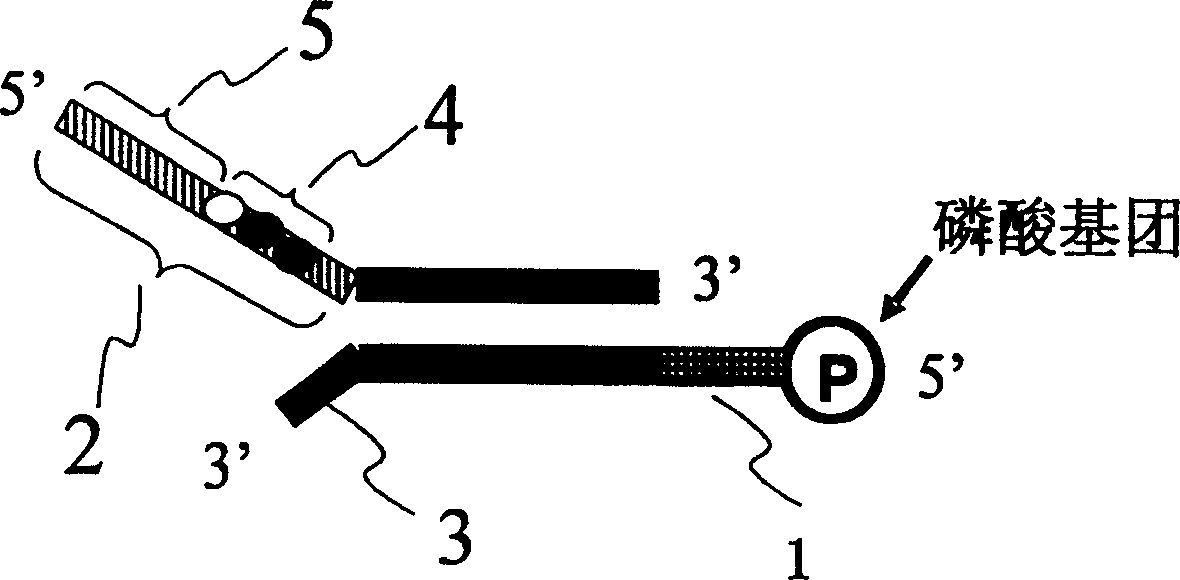
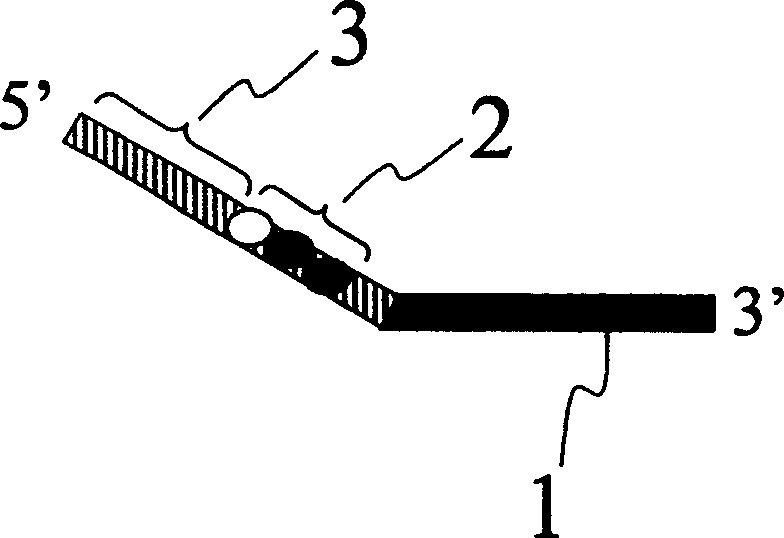
[0025] This example describes the relative expression of P53 gene in three different source tissues by DNA adapter labeling method. Firstly, three different DNA adapters were designed, respectively connected with cDNA fragments digested by restriction endonucleases, and then mixed for PCR amplification.
[0026] 1. Preparation of cDNA samples
[0027](1) Extraction of total RNA: Take 0.1 g each of human normal tissue, brain cancer tissue and liver cancer tissue, add 1 ml Trizol to grind in a tissue grinder, and extract total RNA according to the instructions of Trizol. After the 28s and 18s bands were identified by electrophoresis without degradation, the concentration was determined by ultraviolet absorption spectrometry, and then sterilized DEPC-H 2 O adjusted the final concentration to 1 μg / μl.
[0028] (2) Synthesis of t...
Embodiment 2
[0041] [Example 2] Determination of the difference in the expression of P53 gene between human liver cancer cells and bladder cancer cells.
[0042] This example mainly uses the reverse transcription primer labeling method to determine the difference in the expression of the P53 gene in human liver cancer cells and bladder cancer cells, that is, primers with different sequences are used to reverse transcribe mRNA from different sources, so that the cDNA from each source is labeled DNA fragments with different sequences. And compared with the results of RT-PCR.
[0043] 1. Preparation of Assay Samples
[0044] According to the method of [Example 1], total RNA was extracted from human liver cancer cells and bladder cancer cells respectively. After the quality was verified by electrophoresis, the concentration was measured by ultraviolet light, and then the final concentration was adjusted to 1 μg / μl with DEPC-H20. The mRNA in human liver cancer cells and bladder cancer cells w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com