Method for quick trace synchronous detection of bacteria
A technology for synchronous detection and bacteria, applied in the direction of microbial measurement/inspection, biochemical equipment and methods, etc., can solve problems such as multi-materials, and achieve the effect of synchronous detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
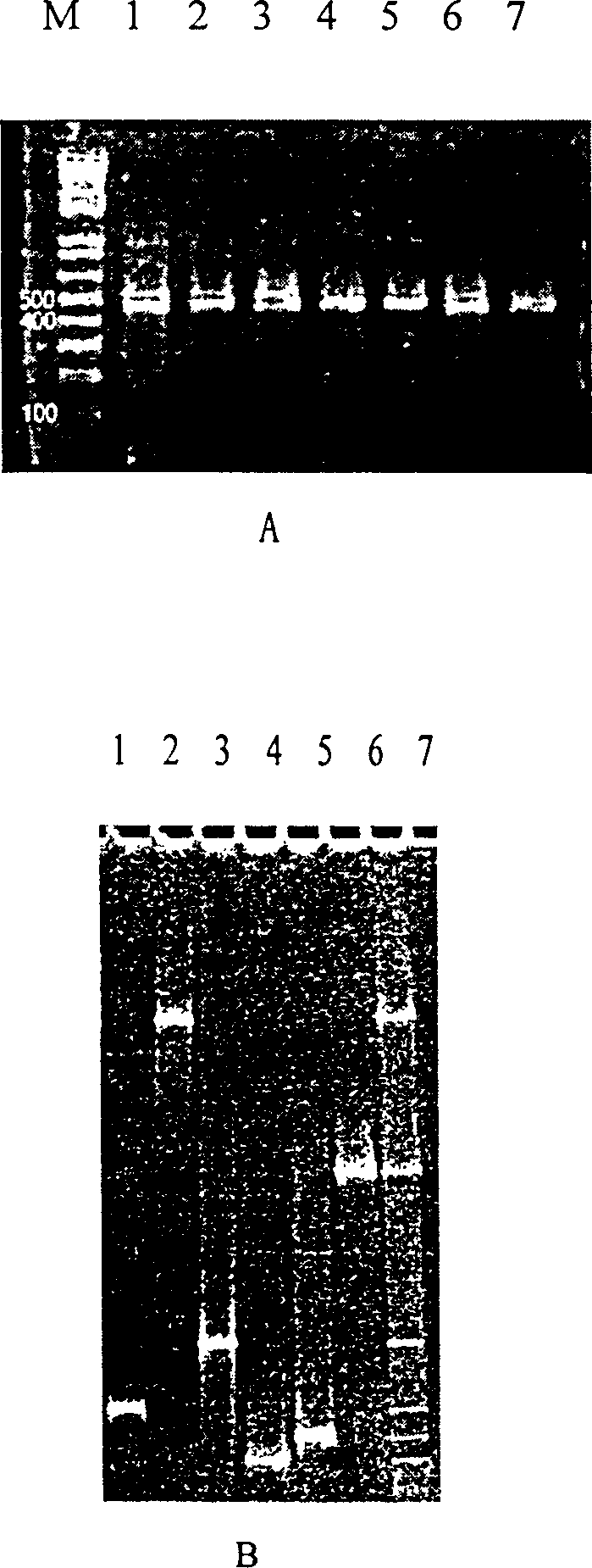
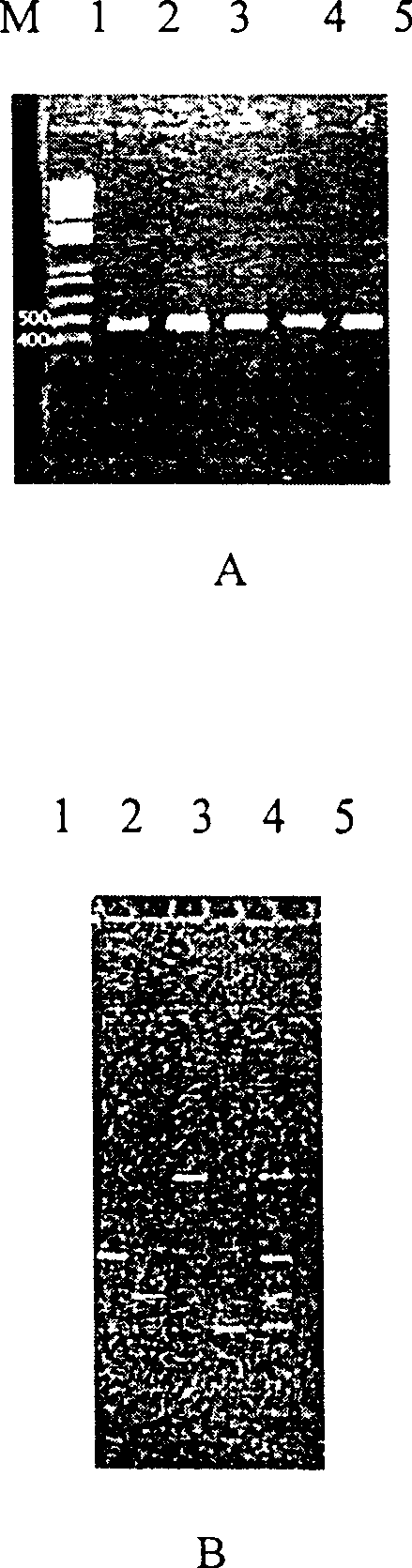
[0014] Use the direct thermal denaturation method to prepare the template, inoculate the bacteria in LB medium, and culture them on a shaker at 28°C for 12-18 hours as seed bacteria, and then inoculate them in the same medium as above at a ratio of 1:50 (bacteria:medium). Incubate on a shaking table at 28-37°C for 12-13 hours; take 0.5ml of the bacterial solution into a sterile 1.5ml centrifuge tube, centrifuge at 3,500 rpm for 20min, discard the supernatant; wash the precipitate with distilled water once, and add 0.8ml of sterile double-distilled water , mix well, boil in 100℃ water bath for 10min; put the centrifuge tube after water bath in 0℃ ice bath to cool, then centrifuge at 12,000rpm for 10min, absorb 15~30μl (25μl PCR reaction system takes 15μl, 50μl PCR reaction system takes 15μl, 50μl PCR reaction system takes 30 μl) supernatant was used as a PCR reaction template.
[0015] Add 2.5 μl 10× buffer, 0.5 μl dNTP (nucleotide mixture, each 10 mM), 0.7 μl MgCl in order 2 ...
Embodiment 2
[0018] Similar to Example 1, the difference is that the template is prepared by thermal denaturation after antibody capture, and antibodies (such as type and group-specific antibodies) that can react with various bacteria are diluted with pH9.6, 0.01mol / L carbonic acid buffer Coat each well of a 96-well enzyme-linked reaction plate (1-10 μg / ml), 50 μl per well, and store in a refrigerator at 4°C overnight; then use pH 7.4, 0.01mol / L phosphate-Tween buffer (PBS- T) Wash the plate 3 times, each time for 3-5 minutes; add 50 μl of 10% calf serum to each well, block at 36-38°C for 1 hour, and shake dry; add 20-40 μl of bacterial liquid culture solution to each well (add 20 μl , 50 μl PCR reaction system plus 40 μl), incubate at 36-38°C for 1-2 hours, wash the plate 5 times with PBS-T washing solution, 3-5 minutes each time. Add 20-40 μl (consistent with the volume of the added bacterial solution) sterile double-distilled water to each well, heat in a boiling water bath for 8-10 min...
Embodiment 3
[0020]Bacterial template preparation: using Pseudomonas fluorescens, Vibrio anguillarum, Vibrio fluvialis, Aeromonas hydrophila, Providencia rettii rettgeri), Aeromonas sobria (Aeromonas sobria), the strains preserved by the applicant. The above-mentioned bacteria were inoculated separately and mixed in LB medium, a total of 7 tubes were cultured on a shaker at 28°C for 12-18 hours, and used as seed bacteria. Then inoculate the above-mentioned same culture medium at a ratio of 1:50 (bacteria:medium), culture on a shaking table at 28°C for 12-13 hours; take 0.5ml of the bacteria solution into a new sterile 1.5ml small centrifuge tube, and run at 3,500 rpm Centrifuge for 20 minutes, discard the supernatant; wash the precipitate with distilled water once, add 0.8ml of sterile double distilled water, mix well, boil in a water bath at 100°C for 10 minutes; put the small centrifuge tube after the water bath in an ice bath at 0°C to cool, Then centrifuge at 12,000 rpm for 10 min, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com