Adenoviral vectors for treating disease
An adenovirus and recombinant adenovirus technology, applied in the field of adenovirus vectors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0108] General method and operation vector
[0109] The methods for constructing and propagating human adenovirus vectors are known in the art, and those of ordinary skill in the art understand how to use these methods in the following examples. Such methods include the methods of operation in the following literature: Hitt, M., et al. Methods of constructing and propagating adenovirus. See: Cell Biology: Laboratory Manual; J. Celis (Ed), Academic Press, N.Y. (1996); Graham, F.L. and Prevec, L. Adenovirus-based expression vectors and recombinant vaccines. In: Vaccines: A new way to solve immunological problems. R.W. Ellis (ed) Butterworth. Pp. 363-390; and Graham, F.L. and Prevec, L. Operation of adenovirus vectors. See: Methods in Molecular Biology, Vol. 7: Gene Transfer and Expression Technology E.J. Murray and J.M. Walker (eds) Humana Press Inc., Clifton, N.J.pp109-128, 1991. The materials and methods described in these documents are used below.
[0110] Adenovirus vector:
[0...
Embodiment 2
[0126] Construction of E3 shuttle vector
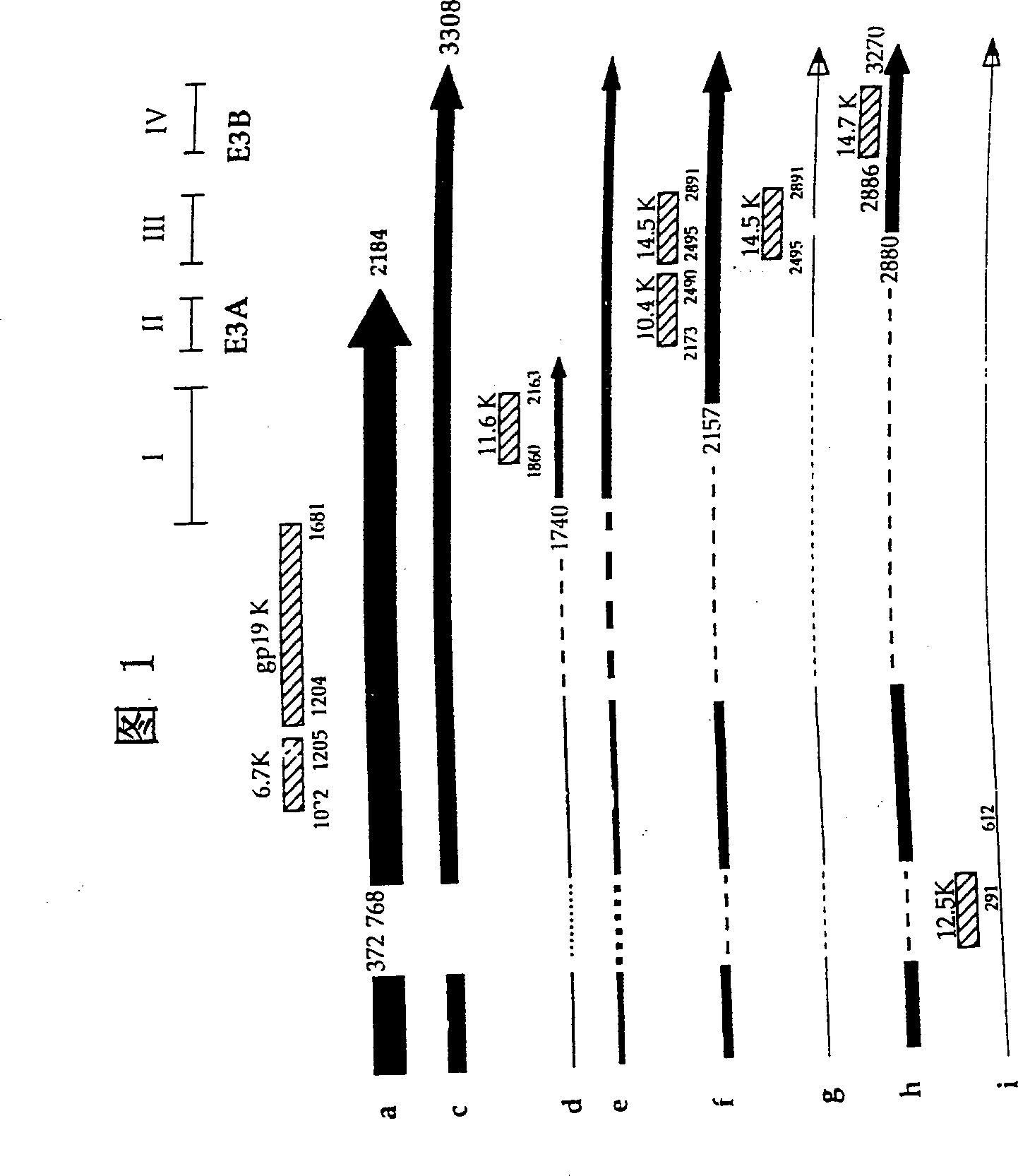
[0127] Using the above vector, the following restriction sites were constructed into the E3 region of adenovirus 5: PacI, ClaI, PmeI, SwaI, BamHI, BstBI, SspI, NheI, and StuI and EcoRV. See Figure 7 for their relative positions in the E3 area. Carefully arrange these restriction sites so that key splicing and polyadenylation signals are not intentionally disrupted (see, Figure 1). In addition, it should be noted that the coding sequence of the protein is: in most cases, do not change the encoded amino acid, when it must be changed, these changes should be conservative. After constructing the virus construct with these changes, compare the splicing, protein level, and function with wild-type Ad5 to confirm the efficacy of the E3 gene in the above shuttle vector.
[0128] Due to the limitation of the location of the engineering site, some mutations have to be carried out sequentially. The sequence of all oligonucleotides used for mutagenesi...
Embodiment 3
[0153] Construction of virus and control
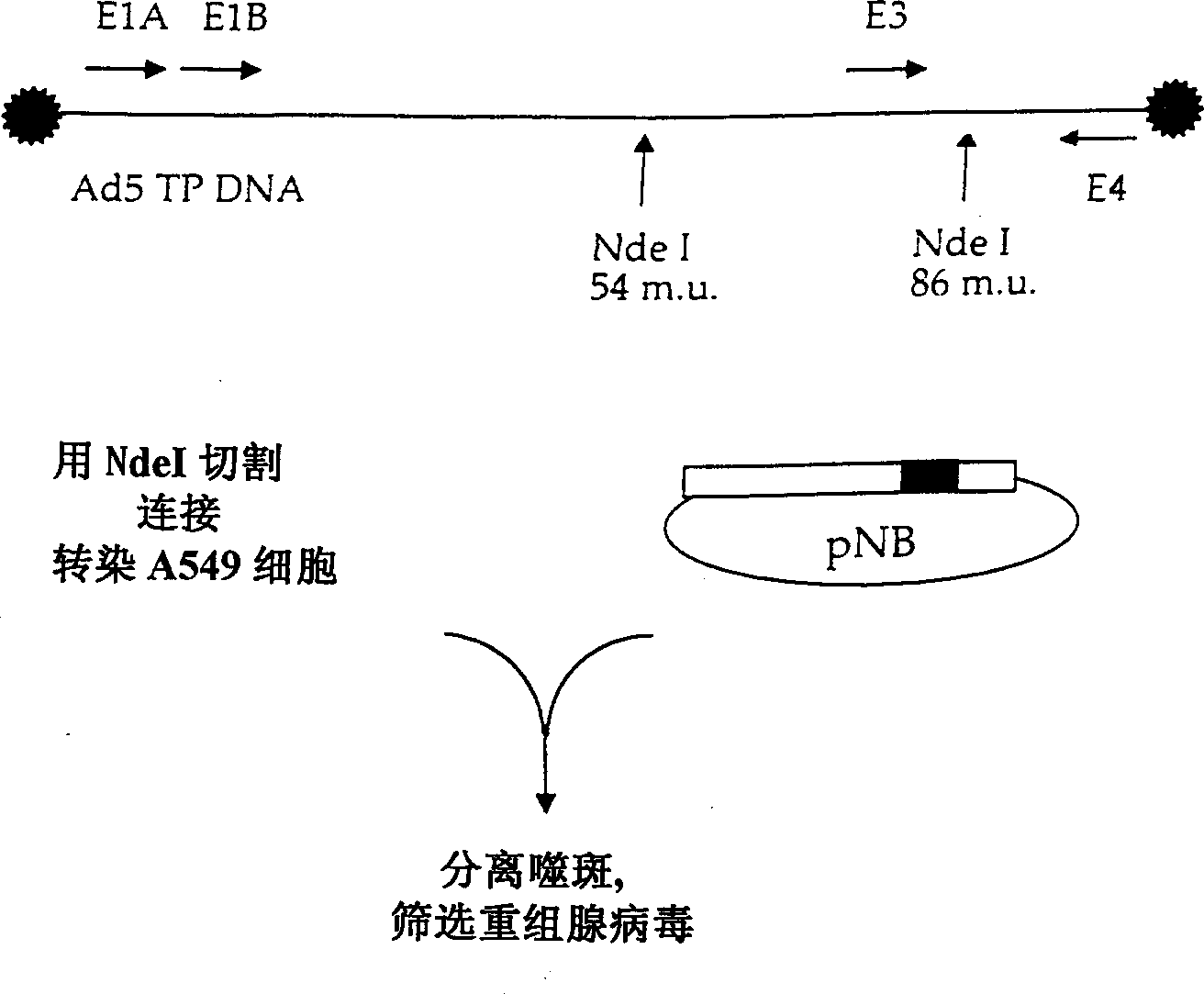
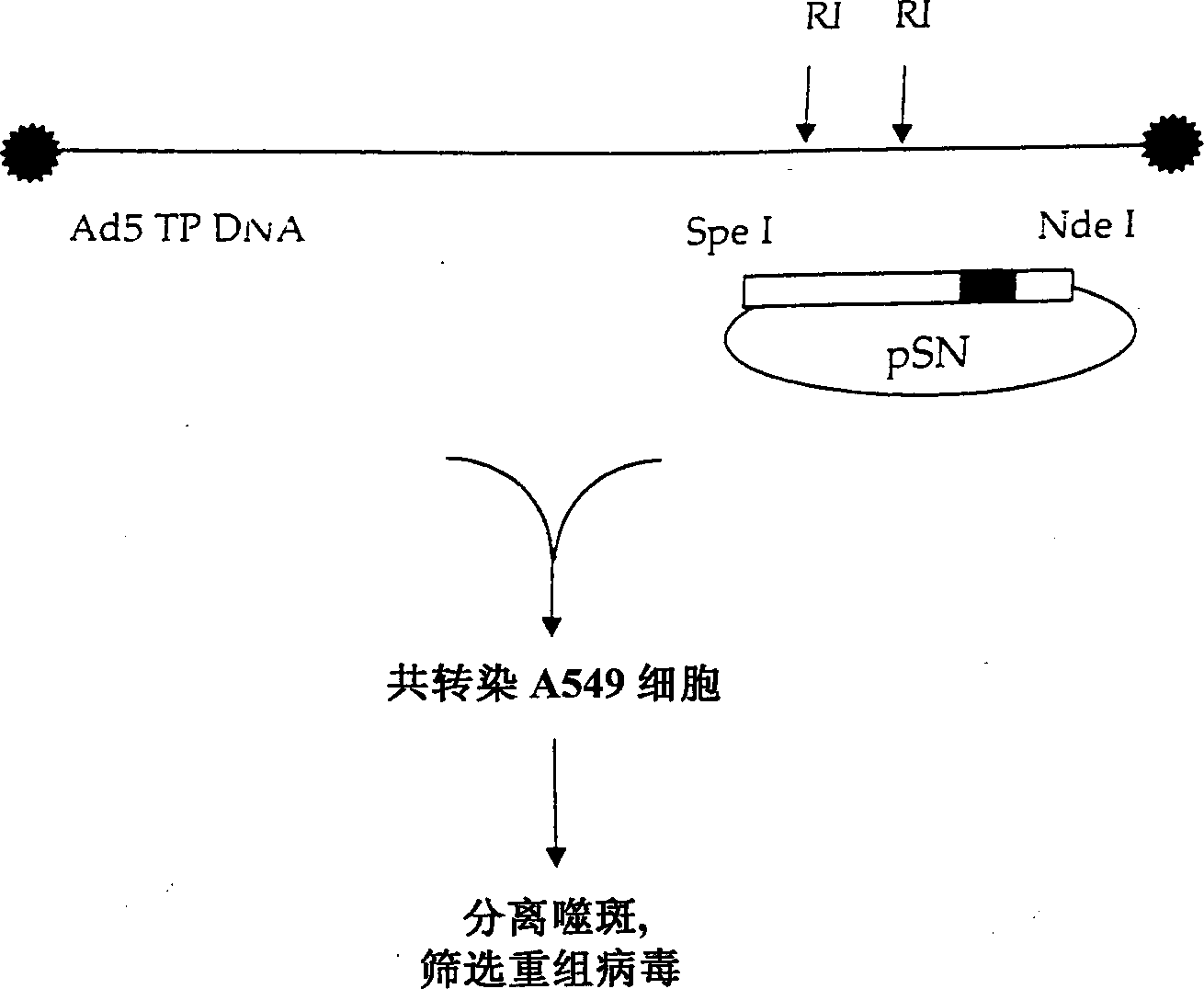
[0154] The last step in virus construction is to insert these altered E3 regions into the pNB plasmid. In order to achieve this operation, the plasmids pG-PPCS and pNB were cut with SpeI. pNB contains two SpeI sites: one in the Ad5 insert and one in pGEMS MCS. The fragment from pNB contains a part of MCS and NdeI19549~SpeI27082. This fragment is inserted into the pG-PPCS plasmid cut by SpeI. The correctness of the orientation was confirmed, and the obtained plasmid was named pNB-PPCS. By inserting the PacI~SwaI region of the smaller plasmid into the larger pNB-PPCS, all the final versions of the E3 shuttle vector were cloned into this plasmid. The resulting plasmids were named pNB-E3SV, pNB-E3SV+V, pNB-E3SV+B, and pNB-E3SV+V+B. Such as figure 2 As shown, these plasmids were then used to prepare corresponding adenoviruses named E3SV, E3SV+V, E3SV+B, and E3SV+V+B. These adenoviruses are deposited in the American Collection of Type Microorg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com