Method for determining bilateral missing sequence of defined DNA sequence utilizing PCR
A DNA sequence and unknown sequence technology, applied in the field of unknown DNA sequences, can solve the problems of inability to obtain regulatory element sequences, consume a lot of manpower and material resources, low abundance and high-level structures, etc., and achieve simple and reliable result analysis, simplify labor and save The effect of time and money
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
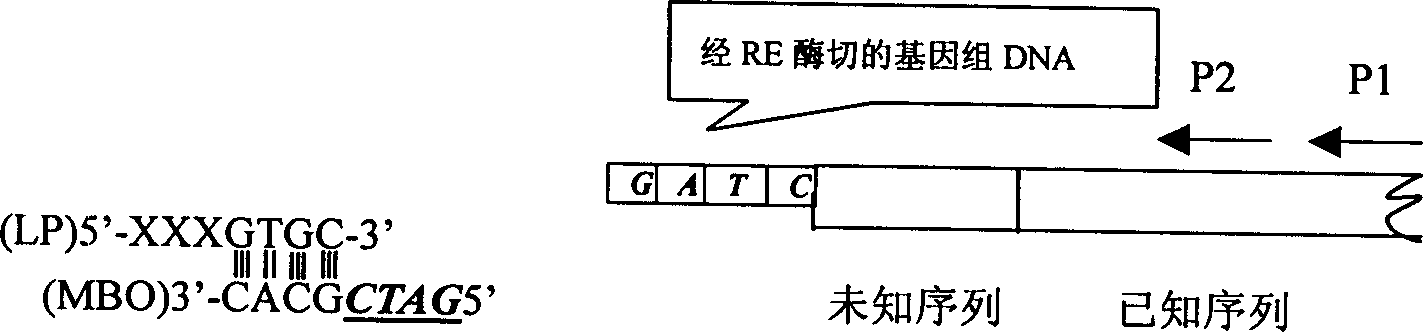
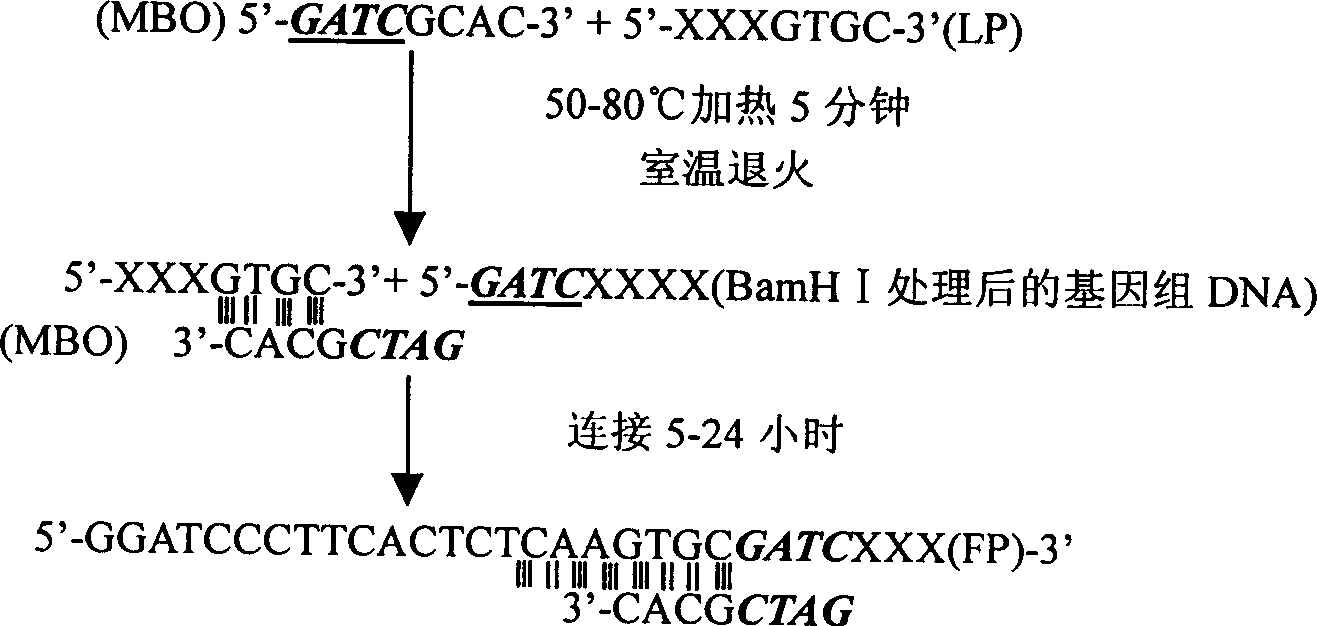
[0030]The amplification of Lip I (Alkaline Lipase I, Penicillium arcum PG37 alkaline lipase I) gene promoter sequence is on the basis of known Lip I DNA sequence, adopts the upstream promoter DNA fragment of the amplified Lip I gene of the present invention (The promoter is located upstream of the gene). Firstly, the 3' end primers P1 and P2 were designed and synthesized according to the known sequence, and P2 was close to the promoter end of LipI. TAQ: 5'-CGGCAC-3' and primer LP 5'-GGATCCCTTCACTTCTCAAGTGC-3' at the junction of the 5' end, use the restriction endonuclease Msp1 whose recognition site is CCGG to digest the PG37 genomic DNA; then TAQ and LP in Heating at 50°C for 5 minutes, annealing at room temperature, and ligation with PG37 genomic DNA digestion product at 25°C for 12 hours, LP will be ligated to the cohesive end of the Msp I digestion site at the 5' end of the unknown sequence to be detected. Using the ligation product as a template and LP and P1 as primers,...
Embodiment 2
[0032] Determination of the Complete Sequence of Archaerhodopsin of Halobacteria halobium Species XZ515 Strain, H.sp.xz515 Strain
[0033] Rhodopsin is a seven-transmembrane protein with light-driven proton pump function. In order to study the reasons for the functional differences between it and other proteins with similar functions, it is necessary to obtain the full-length DNA sequence for comparative biological analysis, in order to expect to obtain the proton pump function of Rhodopsin. Rhodopsin was obtained through purification, and protein sequencing was performed on it. However, due to the closed structure of the protein end, primers designed from protein sequencing could not obtain the full sequence of Rhodopsin DNA, and only a part of the DNA sequence in the middle could be obtained. Its two-terminal sequence can be determined by the present invention. For the N-terminal sequence of Rhodopsin, design primers P1 and P2 through the known DNA sequence in the middle: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com