Method for detecting four kinds of tumor serum proteins
A protein and tumor technology, applied in the field of four common tumor serum proteins of liver cancer, breast cancer, meningioma, and brain glioma detection, which can solve the difficulties of early molecular detection of tumors, evaluation of efficacy and prognosis, recurrence and metastasis monitoring and biological treatment. And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
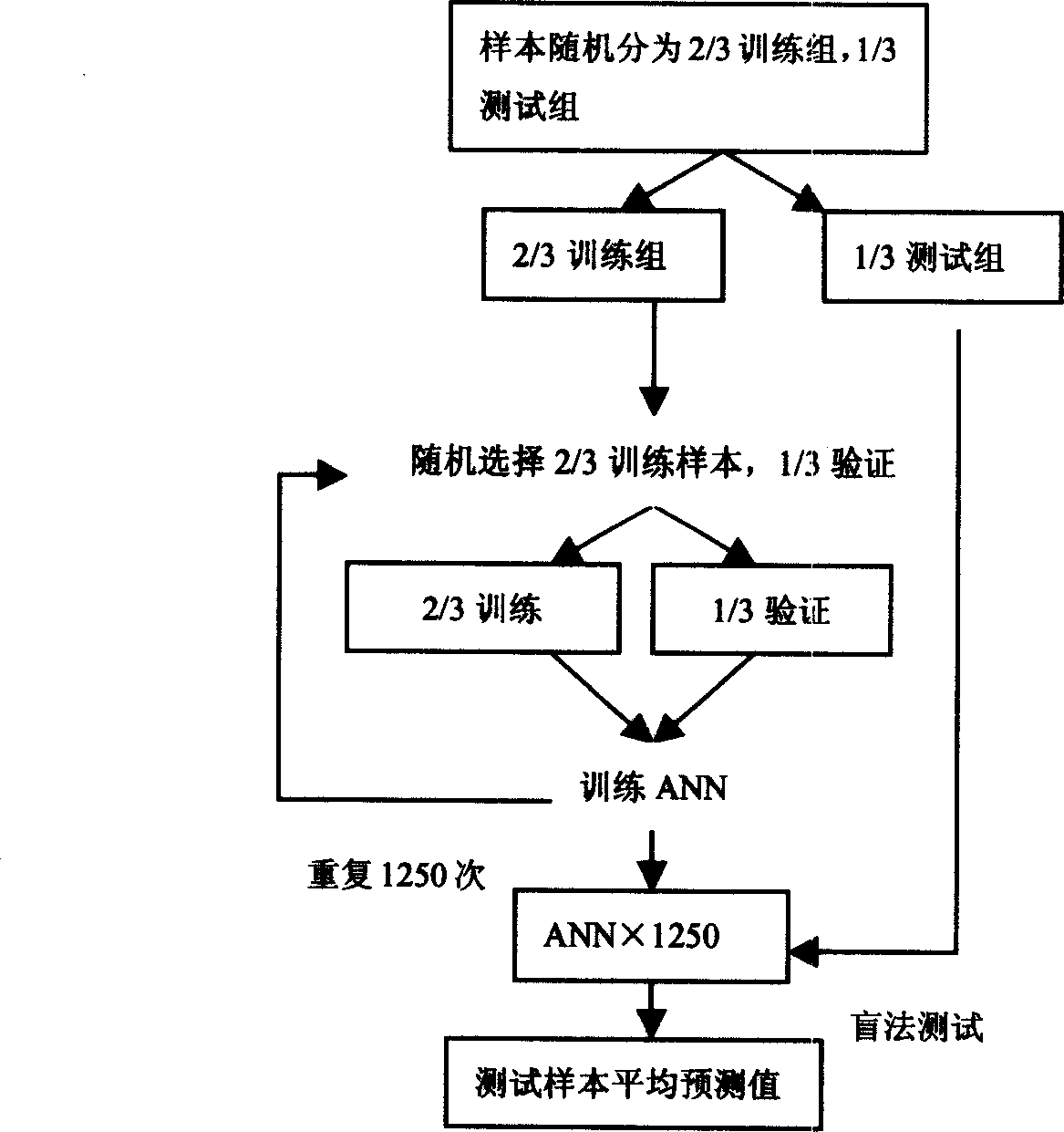
[0022] Embodiment 1 Detection method of the present invention
[0023] 1. Serum samples were thawed in an ice bath and centrifuged at 3000rpm for 5 minutes, and 10ul was diluted with 90ul 0.5% CHAPS (3-cycloethylamine-1-propanesulfonic acid) (pH7.4) solution and then added to Cibacron Blue3GA (white In the protein affinity resin Sigma), react on a shaker at 4°C for 60 minutes, centrifuge at 12000rpm for 5 minutes (4°C), take 50ul of the supernatant solution, and further use 20mM HEPES [N-(2-hydroxyethyl)-piperazine- N'-2 (propanesulfonic acid)] (pH7.4) (H4) / 50mM sodium acetate (pH4.0) (WCX2) was diluted to 200ul, added to the Bioprocessor (96-well sample loaded with H4 / WCX2 chip) (Ciphergen, USA) and vibrated on a shaker for 60 minutes. Shake off the serum specimen, wash the chip with 200ul of 20mM HEPES (pH7.4) (H4) / 50mM sodium acetate (pH4.0) (WCX2) 3 times x 5 minutes, then wash the chip 2 times x 1 minute with deionized water, and dry Afterwards, 0.5ul energy absorbing m...
Embodiment 2
[0036] Example 2 Brain Tumor Detection
Embodiment 3
[0087] Example 3 Breast cancer detection
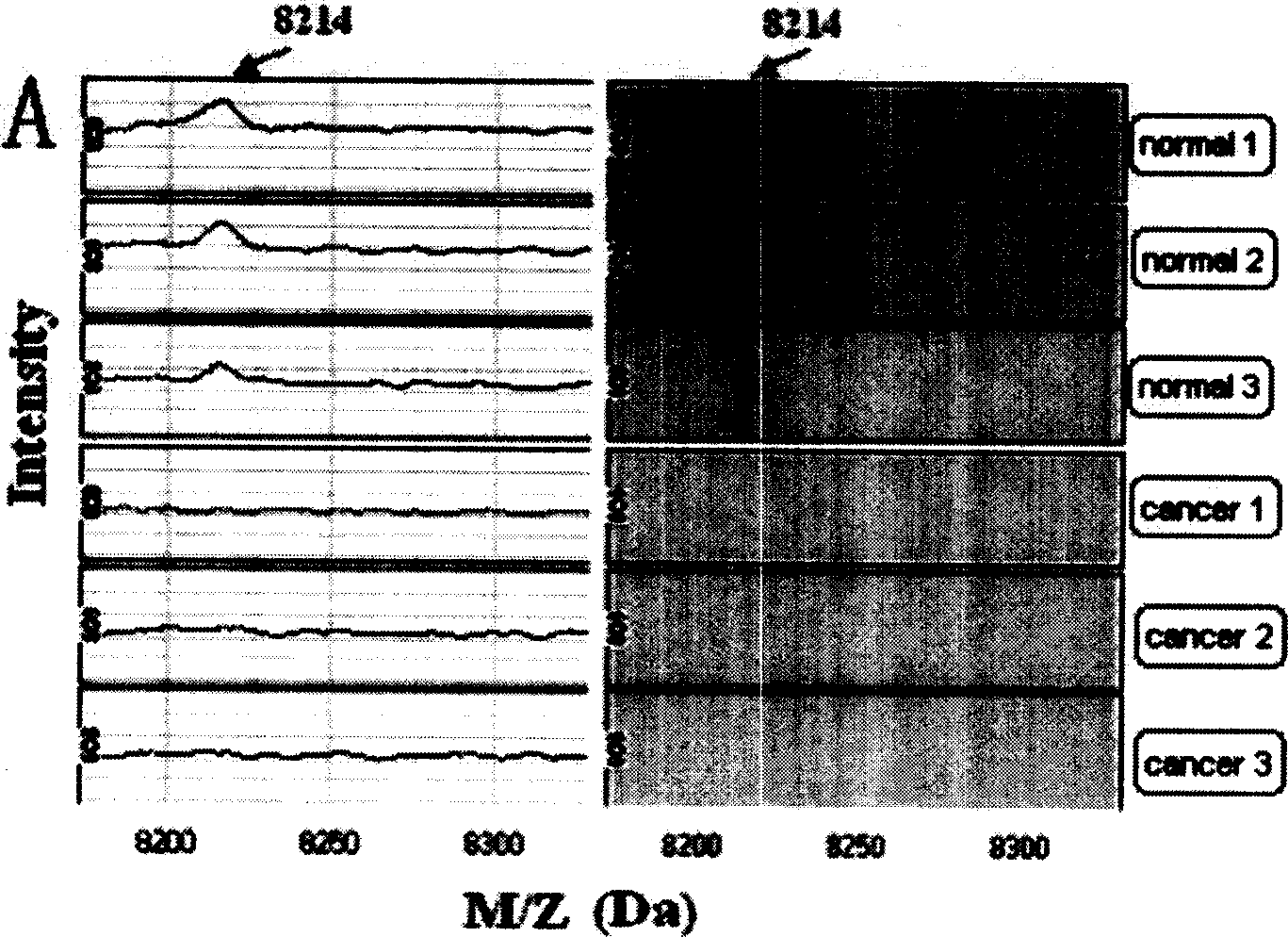
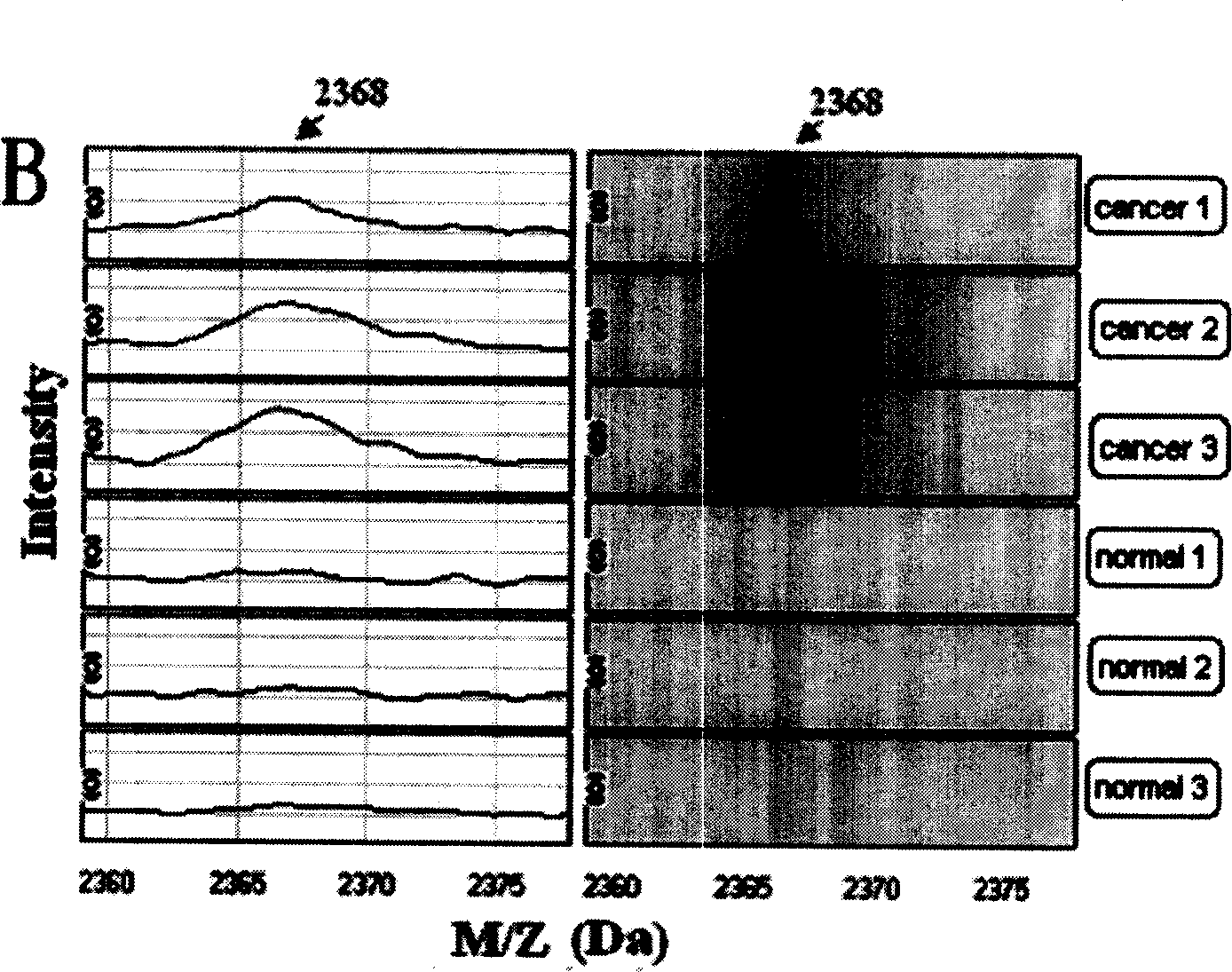
[0088] Using the method in Example 1, the serum protein fingerprints of 49 breast cancer patients, 37 breast cancer patients and 33 healthy people were detected and analyzed. List all the differentially expressed protein peaks obtained in the comparison between breast cancer and healthy human serum protein mass spectrometry data, and use the method of gradually increasing the number of mass-to-charge ratio peaks according to the relative importance to establish multiple comprehensive neural networks and evaluate their prediction success rate, and then The first four mass-to-charge ratio peak combinations with the highest detection accuracy were screened out, and their mass-to-charge ratios were 17.3kDa, 26.3kDa, 5.7kDa and 8.9kDa, respectively. In all 82 specimens, 55 cases (32 cases of breast cancer patients and 23 cases of healthy people) were re-randomly sampled to establish a new training set, and the mass spectrometry data of pro...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com