Systems and methods for predicting specific genetic loci that affect phenotypic traits
A technology for genetic loci and phenotypes, applied in the system field of chromosomal regions, which can solve problems such as limiting the identification of genetic loci
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0146] A second preferred method of making microarrays is to make high density oligonucleotide arrays. Techniques are known for fabricating arrays containing thousands of oligonucleotides complementary to known sequences at defined sites on a surface, which are synthesized in situ using photolithographic techniques (see Fodor et al., 1991 , Science 251:767-773; Lockhart et al., 1996, Nature Biotechnology 14:1675; U.S. Pat. & Bioelectronics 11:687-690). When these methods are used, oligonucleotides of known sequence (for example, 20-mers) can be directly synthesized on the surface of a glass slide or the like. Typically, the fabricated arrays are redundant with several oligonucleotide molecules per RNA. Oligonucleotide probes can be selected to detect alternatively spliced mRNAs.
[0147] Other methods can also be used to make microarrays, for example by labeling (Maskos and Southern, 1992, Nuc. Acids. Res. 20: 1679-1684). In general, any array type can be used, such as d...
Embodiment 4
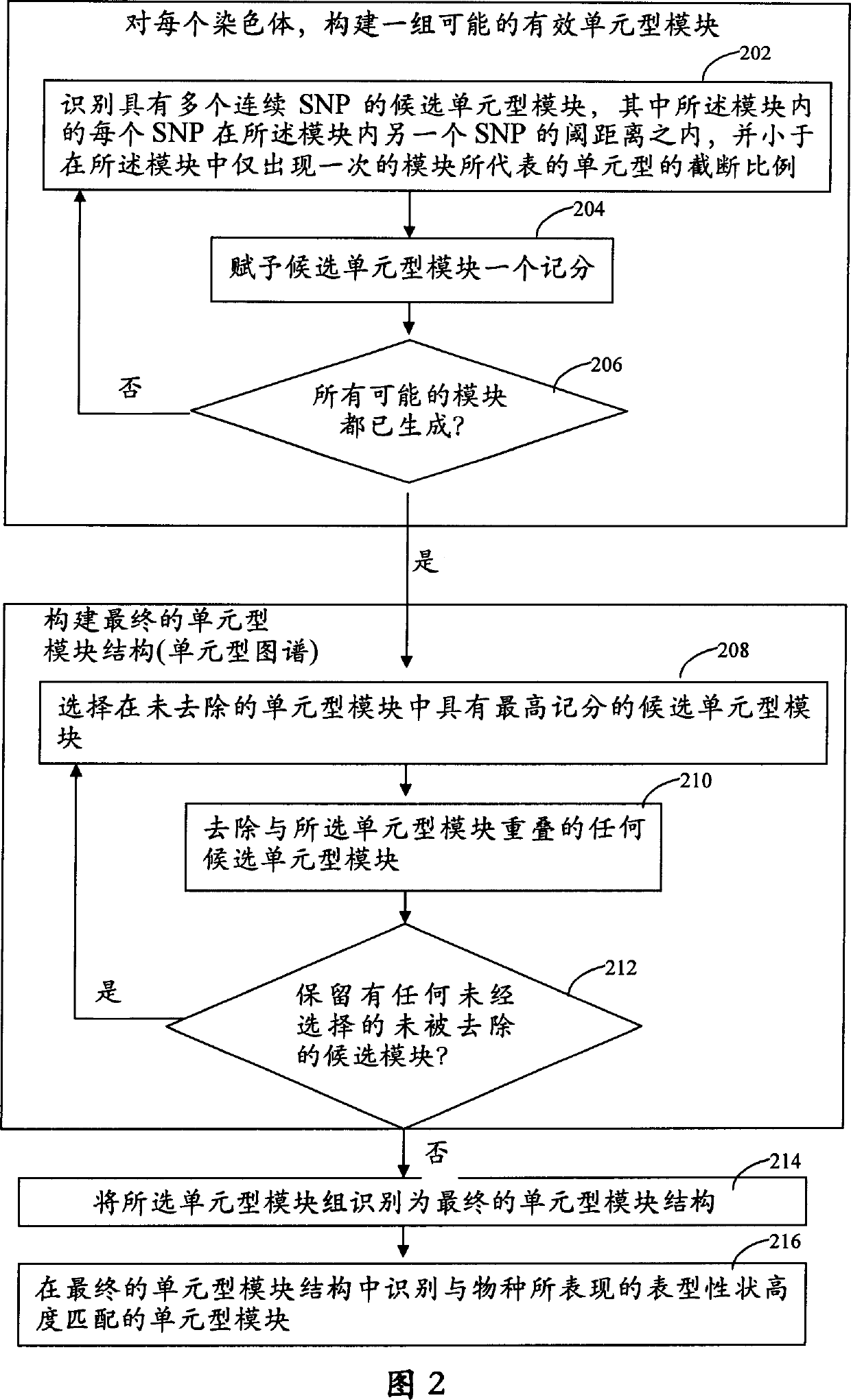
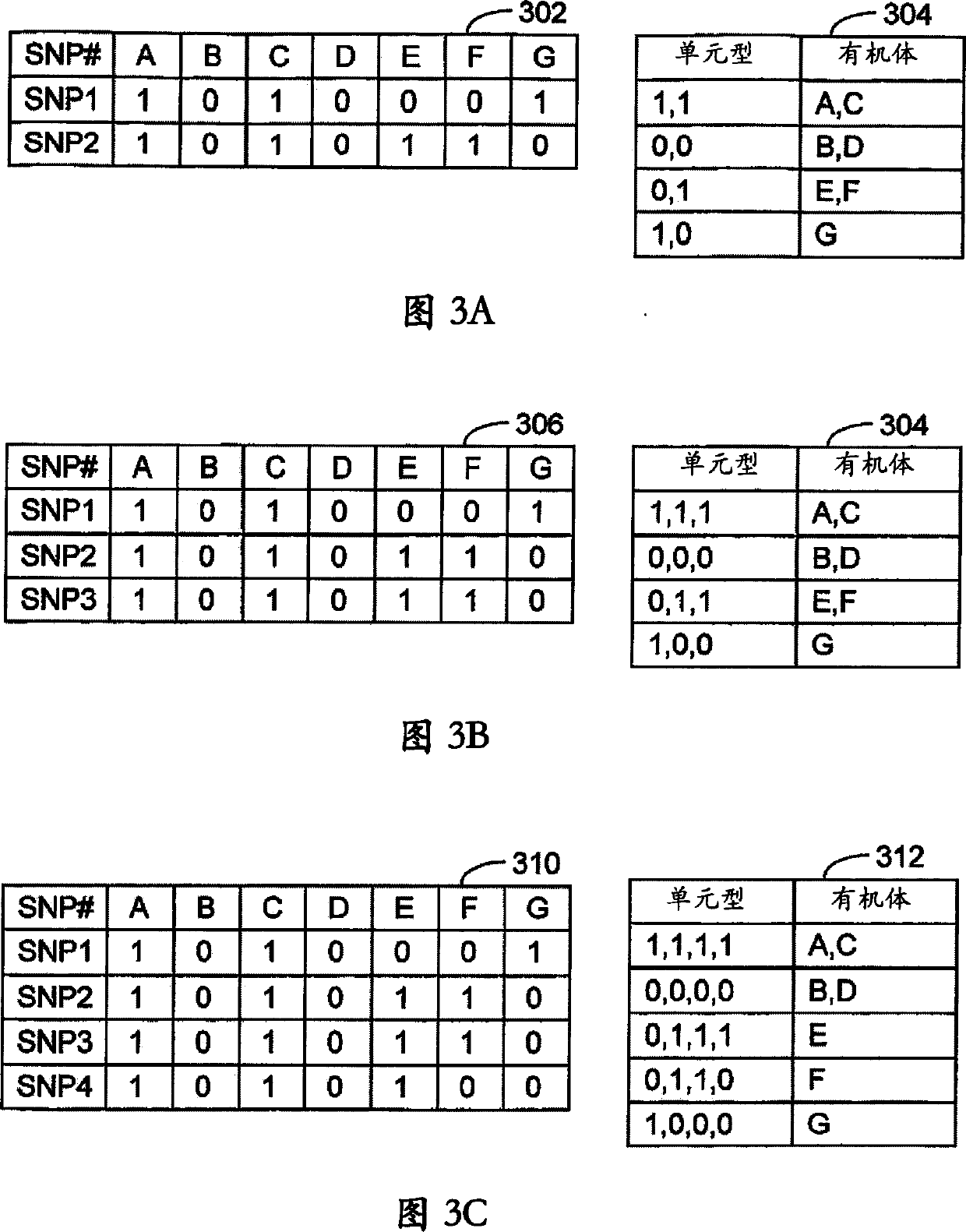
[0169] In Example 1, the characteristics of the haplotype blocks generated using the technique disclosed in FIG. 2 as a function of the number of lines (organisms) present in the genotype database 52 are presented. In Example 2, the systems and methods of the invention were used to correlate phenotypic data obtained from inbred mouse strains with haplotype blocks. In Example 3, the systems and methods of the present invention were used to construct biological pathways. In Example 4, the systems and methods of the present invention were used to determine which chromosomal regions responded to perturbations.
[0170] 1.10.1 Embodiment 1
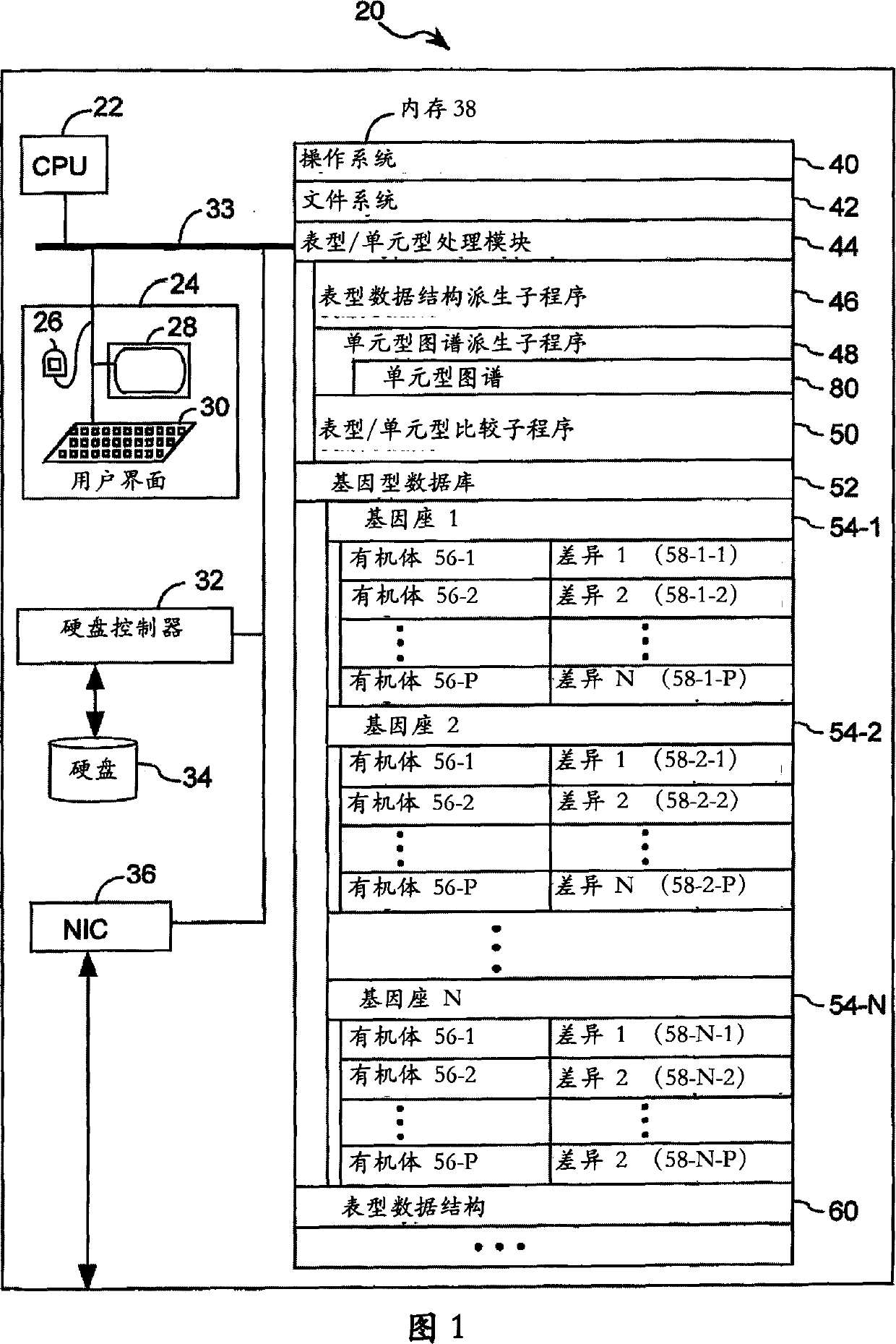
[0171] An exemplary genotype database 52 used in this example was obtained from (http:\\mouseSNP.Roche.com). An automated high-throughput method of resequencing targeted genomic regions to discover SNPs and characterize alleles, see Grupe et al., 2001, Science 292, 1915-1918. The genomic regions analyzed were all within known biologically im...
Embodiment 2
[0184]U.S. Patent Application 09 / 737,918, filed December 15, 2000, entitled "Systems and Methods for Predicting Chromosomal Regions that Control Phenotypic Traits," and entitled "Predictively Controlling Chromosomal Regions of System and Method for Chromosomal Regions," U.S. Patent Application No. 10 / 015,167, which predicts in silico by correlating phenotypic data obtained from inbred mouse strains with the extent to which alleles are shared within genomic regions. Chromosomal regions that regulate complex traits. It can be determined whether comparing complex phenotypes to the haplotype map of the mouse genome is a better method for in silico analysis of mouse phenotypic traits than the methods disclosed in U.S. Patent Application 09 / 737,918 and U.S. Patent Application 10 / 015,167 method. For each haplotype block within the haplotype map, correlations were calculated by calculating the negative logarithm of the ratio of the average phenotypic difference within haplotype group...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com