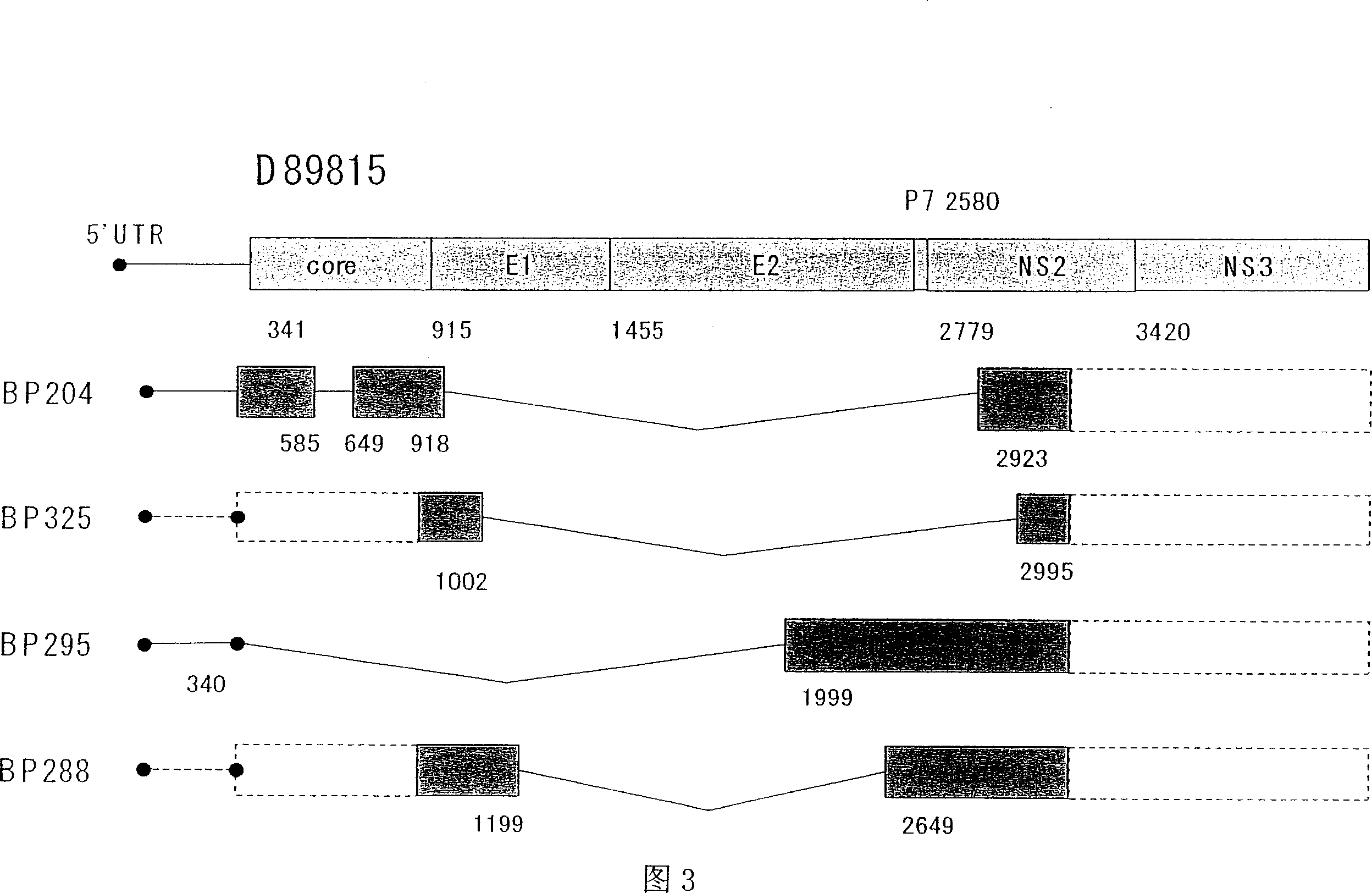
HCV RNA having novel sequence
A hepatitis C virus and gene technology, applied in the direction of viral peptide, virus/phage, recombinant DNA technology, etc., can solve the problem that the hypothesis cannot be verified, and it is not used to prove the HCV infection and proliferation model.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
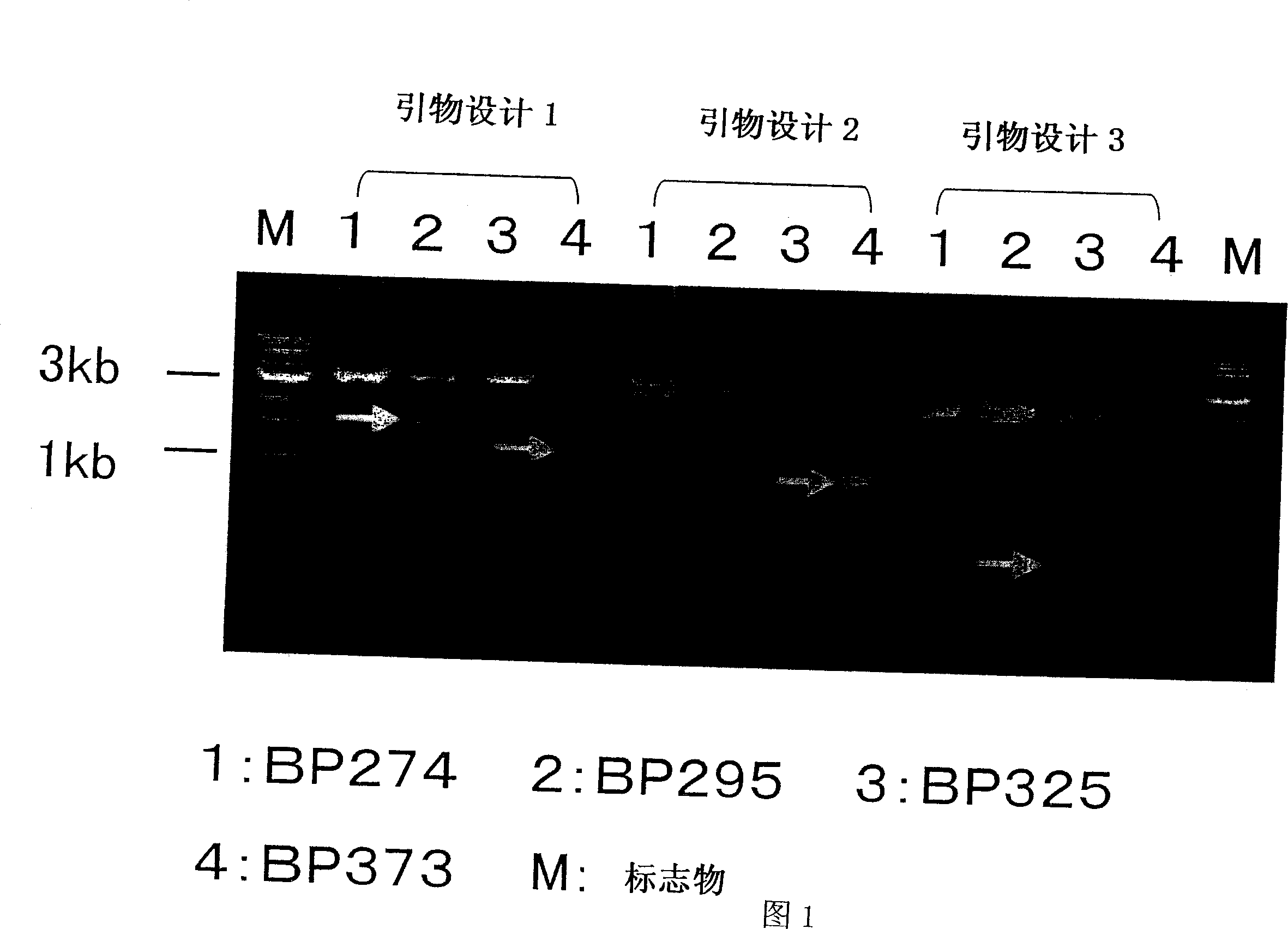
[0110] Example 1. Separation and decoding of shortened sequence
[0111] Slice the patient’s liver with BP207 (0.5mm×1mm) in 100μl of RIPA buffer (20mM TriS-HCl [pH7.5], 150mM NaCl, 1% NP40, 0.1% deoxycholic acid, complete protease inhibitor mixture [Roche diagnostics Corporation]), centrifuged at 10krpm for 5 minutes, and then recovered the supernatant. Use a high-purity viral nucleic acid kit (Roche diagnostics corporation) to purify nucleic acid from the extract according to the method recommended by the manufacturer. HC9405R-1b primer was added to the purified nucleic acid, and MMLV reverse transcriptase (Invitrogen) was used to perform reverse transcription reaction at 42°C for 1 hour under the conditions recommended by the manufacturer to obtain cDNA.
[0112] RNaseH (Invitrogen) was added to the reaction solution and reacted at 37°C for 30 minutes to decompose RNA. Using a part of the reaction solution, in the presence of HC-LongAl primers and T7-HC9313R primers, use Klen...
Embodiment 2
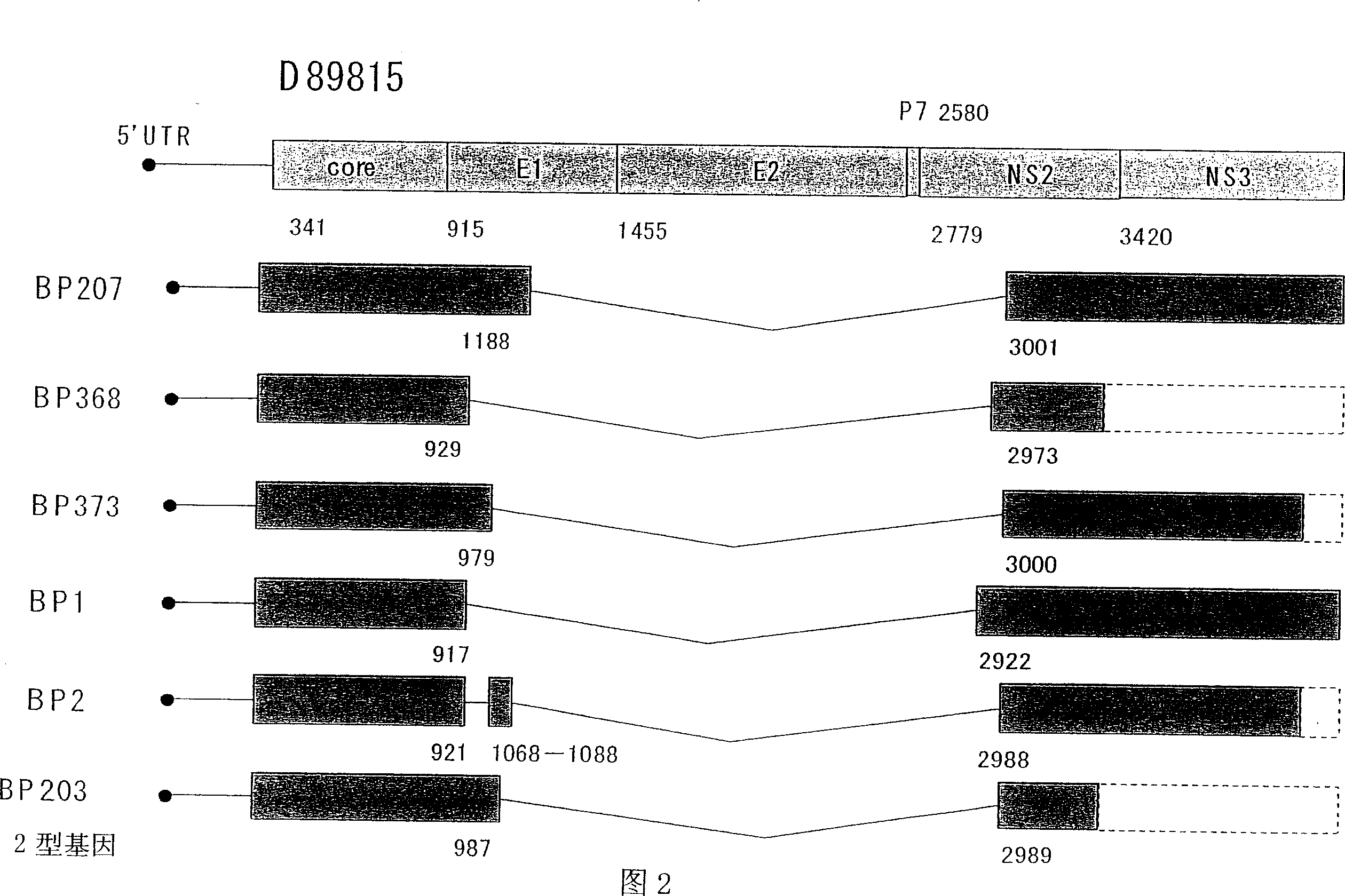
[0145] Example 2. Isolation and analysis of cDNA of TF HCV genomic RNA from patients with Lyceum code
[0146] Next, 23 specimens from liver biopsy of liver with chronic active hepatitis and tissue specimens BP1, BP2, BP3 of liver cancer patients during surgery, and 3 specimens were tried to detect HCV-RNA of TF. Out.
[0147] RNA extraction
[0148] The RNA was extracted based on the rules of "Isolation of RNA from Micro Samples" of ISOGEN (Japan GENE Co., Ltd.), which is a test drug for RNA extraction. Add 0.8ml of ISOGEN to the patient's liver slice with a size of about 0.5mm×1mm, use a 1ml needle tip to decompose the tissue piece, and then pulverize by suction. Leave it in the greenhouse for 5 minutes, add 0.2ml of chloroform, stir vigorously for 30 seconds, and leave it at 4°C for 5 minutes. Centrifuge at 12,000 g in a small refrigerated centrifuge for 15 minutes at 4°C to recover the water phase.
[0149] Add about 5ug of yeast tRNA, add 0.8ml of isoproterenol, and pla...
Embodiment 3
[0213] Example 3. Preparation of HCV RNA replicon
[0214]By inserting a linking fragment obtained by heat-treating XhoX-Xba-s oligomer and XhoX-Xba-as oligomer between the XhoI and XbaI restriction sites of pBluescriptIISK(+), pBSIISK(+)△XX was constructed. In addition, pLV207-0007 was provided by PCR with Sbf_H1 primer and Cla_as primer, and a fragment of about 0.7kb was amplified, and then cloned by pGEM-T Easy to obtain pLVC-0007Sbf. By inserting NotI and ClaI between the NotI and XbaI restriction sites of pBSIISK(+)△XX, the fragment of about 0.7kb can be obtained by digesting pLVC-0007Sbf with ClaI and XbaI, and pLVC-ClaXba7.2K can be digested with ClaI and XbaI. The resulting fragment of about 7.2 kb yielded pSbf-LV207TF.
[0215] On the other hand, T7-HC9313b primer was added to HCV antibody-positive serum and RNA purified from G14, and cDNA was synthesized by SuperscriptII reverse transcriptase (Invitrogen) using the method recommended by the manufacturer. Using a part o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com