Process to study changes in gene expression in granulocytic cells
a technology of granulocytic cells and gene expression, applied in the field of process to study changes in gene expression in granulocytic cells, can solve the problems of eosinophil cationic, systematic approach to assess transcriptional response, increased blood flow (hyperemia) and edema
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Production of Gene Expression Profiles Generated from cDNAs Made with RNA Isolated from Neutrophils Exposed to Virulent and Avirulent Bacteria.
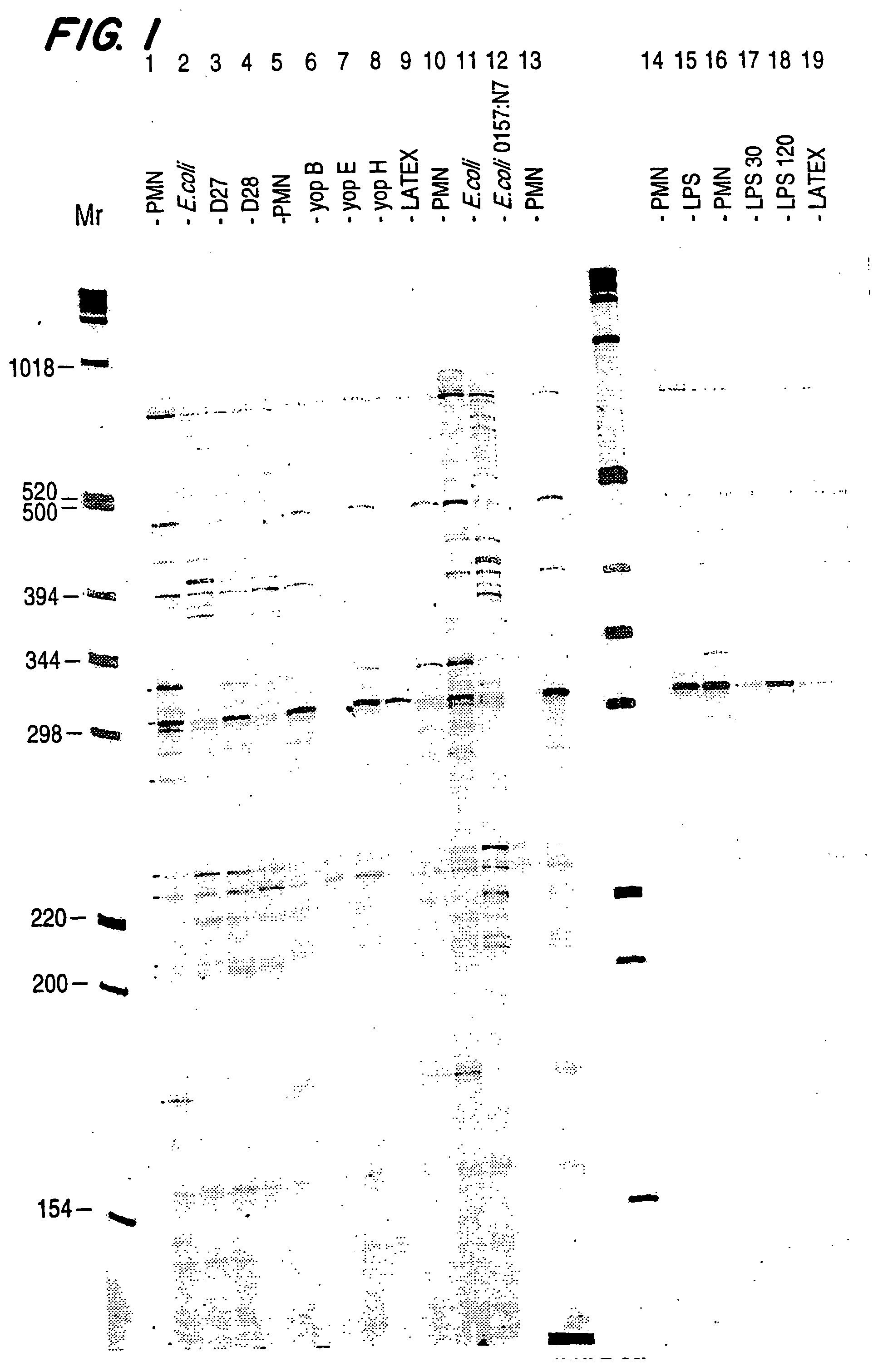
[0078] Expression profiles of RNA expression levels from neutrophils exposed to various bacteria offer a powerful means of identifying genes that are specifically regulated in response to bacterial infection. As an example, the production of expression profiles from neutrophils exposed to virulent and avirulent E. coli and Y. pestis allow the identification of neutrophil genes that are specifically regulated in response to bacterial infection.
[0079] Neutrophils were isolated from normal donor peripheral blood following the LPS-free method. Peripheral blood was isolated using a butterfly needle and a syringe containing 5 cc ACD, 5 cc of 6% Dextran (in normal saline). After 30 minutes of settling, plasma was collected and HBSS (without Ca++ or Mg++) was added to a total volume of 40 ml. The plasma was centrifuged (1500 rpm, for 15 m at 4° C....
example 2
Production of Gene Expression Profiles Generated from cDNAs Made with RNA Isolated from Neutrophils Exposed to Virulent and Avirulent Bacteria and Neutrophils Exposed to Cytokines.
[0088] Neutrophils were isolated from normal donor peripheral blood following the LPS-free method as set forth in Example 1.
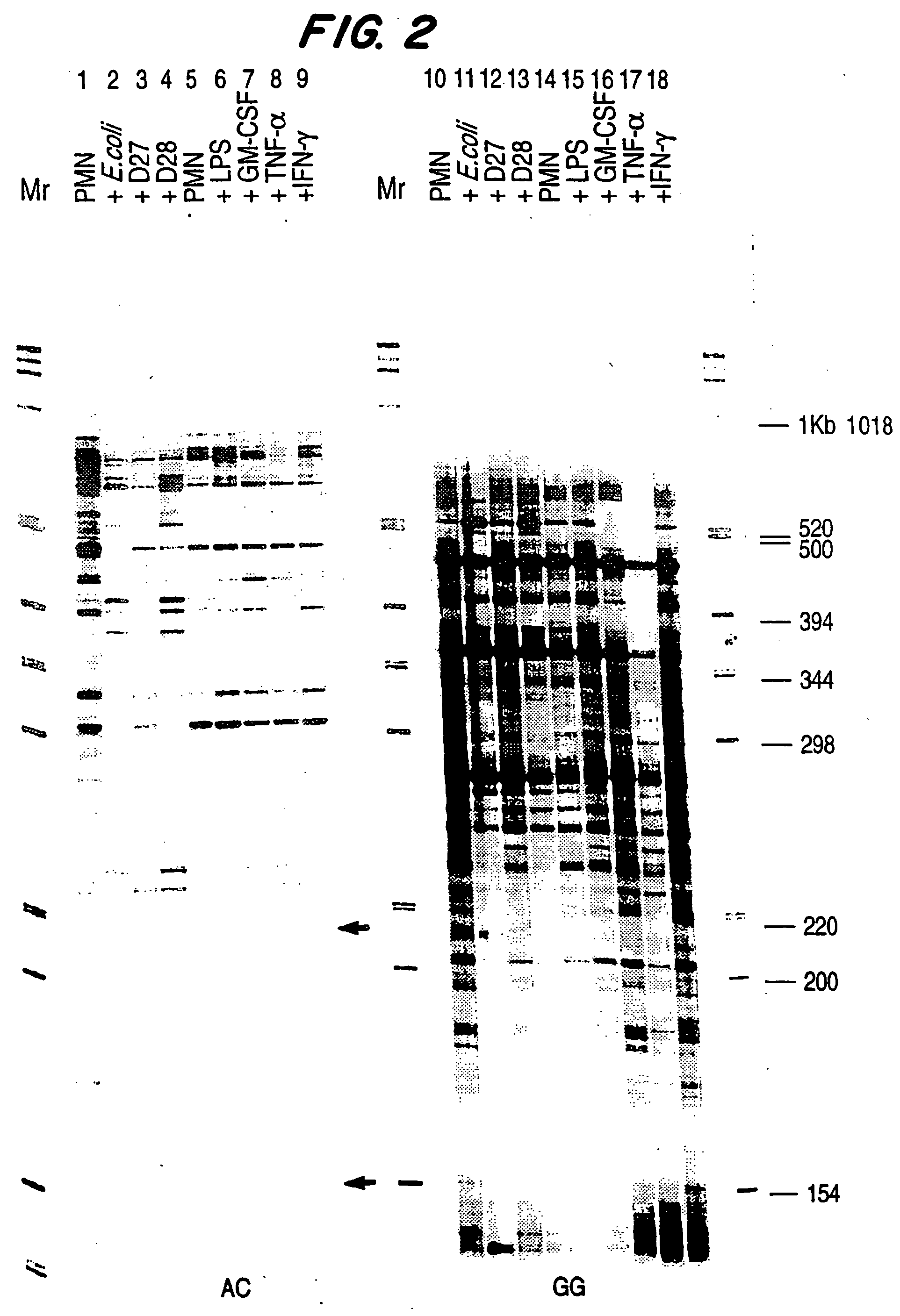
[0089] Neutrophils were incubated with virulent and avirulent E. coli or Y. pestis, LPS at 1 ng / ml, GM-CSF at 100 units / ml, TNFα at 1000 units / ml, or γIFN at 100 units / ml. The bacterial cells, LPS or cytokines were added to approximately 3.38×108 cells in 100 ml of RPMI containing 6% H1 autologous serum. Incubation proceeded for 2 to 4 hours, preferably 2 hours, with gentle rotation in disposable polycarbonate Erlenmeyer flasks at 37° C. After incubation, the cells were spun down and washed once with HBSS.
[0090] After incubation of the neutrophils, RNA was extracted and the cDNA profiles prepared as described in Example 1. FIG. 2 is an autoradiogram of the expression profiles gene...
example 3
Production of Gene Expression Profiles Generated from cDNAs Made with RNA Isolated from Neutrophils Exposed to Bacteria Using all 12 Possible Anchoring Oligo d(T) n1,n2.
[0092] Neutrophils were isolated from normal donor peripheral blood following the LPS-free method.
[0093] Neutrophils were incubated with E. coli or Y. pestis.
[0094] After incubation of the neutrophils, RNA was extracted and the cDNA profiles prepared as described in Example 1. FIG. 3 is an autoradiogram of the expression profiles generated from cDNAs made with RNA isolated from control (untreated) neutrophils (lane 1), neutrophils incubated with avirulent E. coli K12 (lane 2), virulent Y. pestis (lane 3), avirulent Y. pestis (lane 4). The anchoring oligo d(T)18 n1 and n2 positions are indicated at the top of the figure. The cDNAs were digested with BglII.
[0095]FIG. 4 represents a summary of genes which are differentially expressed in neutrophils upon exposure to virulent and avirulent E. coli and Y. pestis. Expr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| total volume | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com