Screening method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
CEACAM-1 Expression in Adenomas of the Colon
[0109] Freshly-derived samples of adenomas from patients were investigated for the expression of different CEACAM genes (Nollau P, et al., (1997), Cancer Res. 57, 2354-2357). For this purpose, total RNA was isolated from the samples through a conventional method and analyzed using Northern Blot (Neumaier M. et al., PNAS, (1993), 90 (22), 10744-10748).
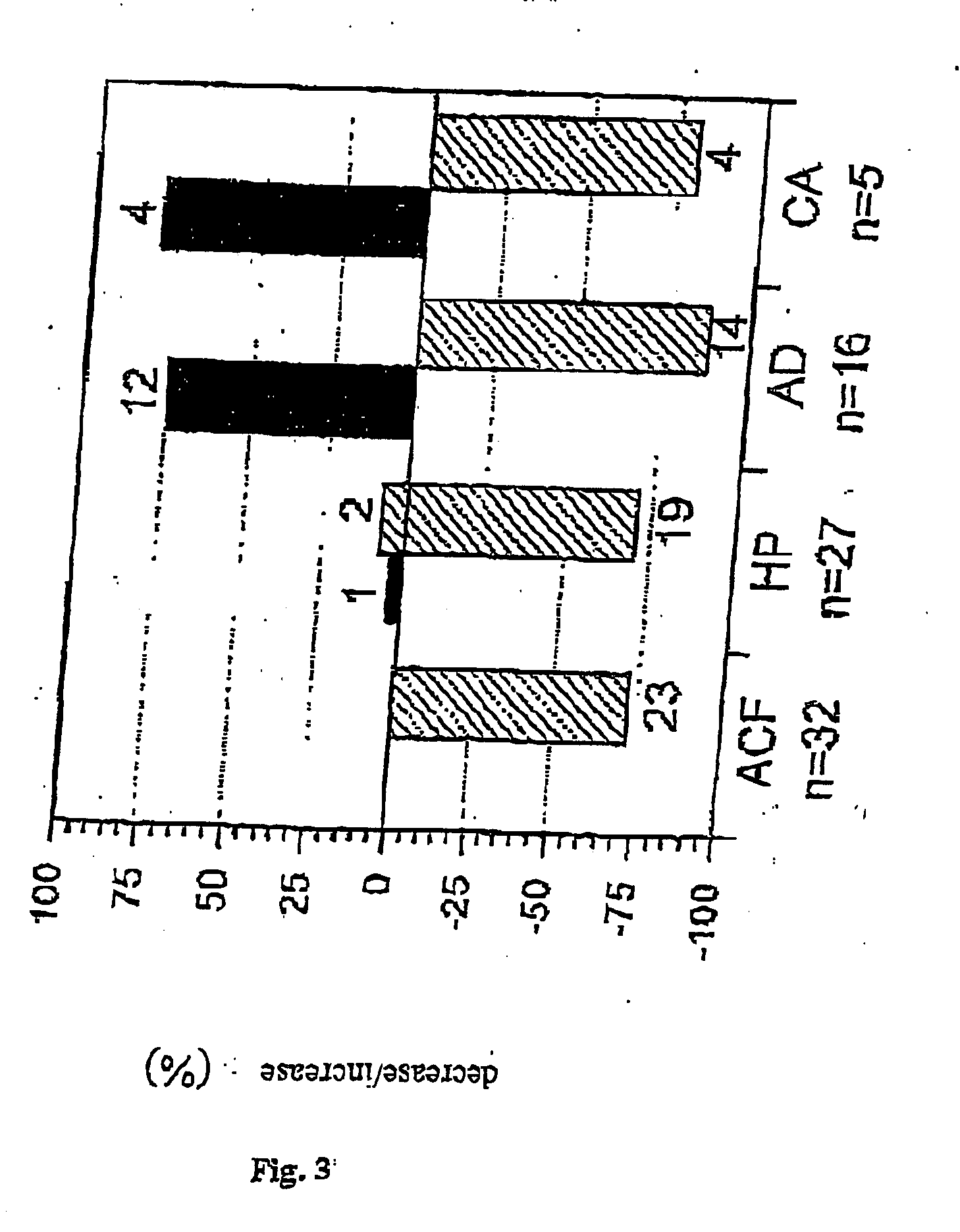
[0110] The Northern Blots were evaluated using quantitative densitometry and image analysis and the alterations in the expression of CEACAMs compared to the corresponding normal tissue of the same patient (matched-pair analysis). The results for the adenomas are presented in Table 1 below.
TABLE 1Sample No.CEACAM1 expression 8BIØ 9BII((+))13BIØ13BII((+))14BIØ14BII((+))15BI((+))15BII(+)18BI((+))19BII((+))23BIØ23BIIØ24BI((+))25BIØ25BIIØ30B(+)31B(+)32B((+))
Ø = Specific CEACAM1 RNA no longer detectable
(+) = 60% loss in expression
((+)) = >80% loss in expression
example 2
Immunohistochemical Demonstration of the Change in CEACAM1 Expression in Different Precursor Lesions
[0111] Normal colon mucosa tissue was freshly obtained from surgical interventions. The material was fixed in 4% formaldehyde / PBS at 4° C. for 1 to 4 hours using a standard protocol. The samples were stained with 0.2% methylene blue (Sigma, Taufkirchen, Germany) for 3 minutes and investigated for the presence of ACF using a magnifying glass. Samples with more than 7 foci were used for further investigations. Samples of hyperplasic polyps, adenomas and carcinomas were obtained from the Pathology Institute of the University Hospital in Hamburg for comparison purposes.
[0112] 5 μm sections were derived from the tissue samples embedded in paraffin by a standard technique. The paraffin was removed from the sections on microscope slides and the sections washed with PBS. The sections were then pre-treated in a microwave for 10 minutes at 650 W in 10 mM citrate buffer (pH 6.0) or for 3 minut...
example 3
Significance of the Expression of CEA (CEACAM-5) for Apoptosis Sensitivity in HT29 Cells
[0123] Ribozyme-regulated mRNA expression is used to investigate the influence of CEA protein expression on the susceptibility of the cells to apoptosis. HT29 colon cancer cells (obtained from ATCC, Rockville, USA) were transfected with CEA-targeted hammerhead ribozyme expression vector.
[0124] The vector was derived by a method analogous to that of Schulte et al., PNAS 1996, 93, pages 14759ff and Juhl, H., et al, 1997, JBC 272, pages 29482ff.
[0125] The following oligonucleotides were used as ribozyme sense and antisense nucleotides.
5′-agcttTGCTCTTCTGATGAGTCCGTTAGGACGAAACTATGGAgggcc-3′ (sense)(SEQ ID NO. 1)and5′-cTCCATAGTTTCGTCCTAACGGACTCATCAGAAGAGCAa-3′ (antisense)(SEQ ID NO. 2)
[0126] These were hybridized and ligated in the pTET vector, as described by Schulte et al,, PNAS 1996, 93, pages 14759ff. The plasmid obtained, pTFT / Rz2113, contains CEA-specific flanking regions at the 5 ′ end (7 nu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com