Immunostimulatory nucleic acid molecules
a nucleic acid and immunosuppression technology, applied in the field of oligonucleotides, can solve problems such as poly(i,c), and achieve the effect of increasing the sensitivity of chronic leukemia cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Effects of ODNs on B Cell Total RNA Synthesis and Cell Cycle
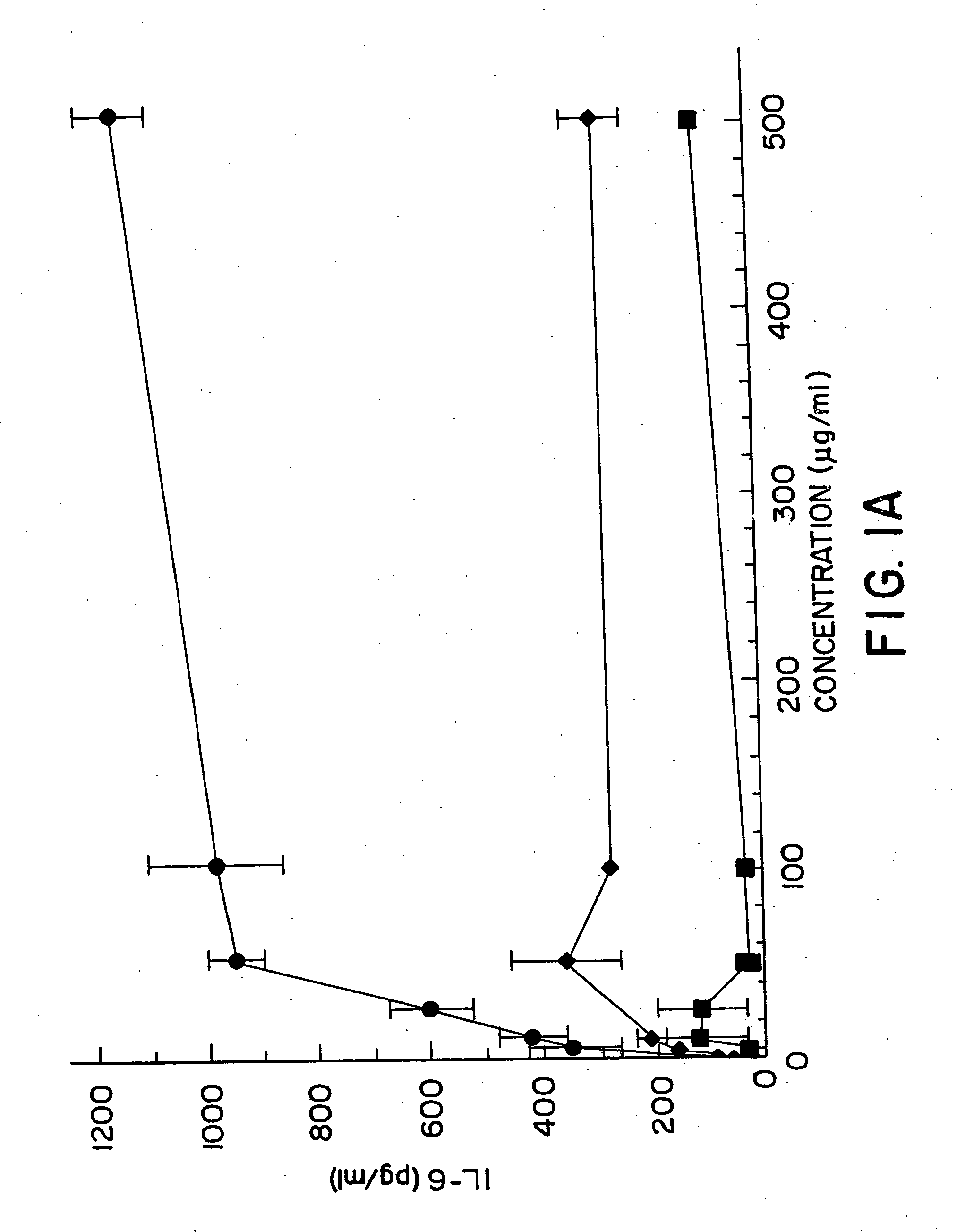
[0152] B cells were purified from spleens obtained from 6-12 week old specific pathogen free DBA / 2 or BXSB mice (bred in the University of Iowa animal care facility; no substantial strain differences were noted) that were depleted of T cells with anti-Thy-1.2 and complement and centrifugation over lymphocyte M (Cedarlane Laboratories, Hornby, Ontario, Canada) (“B cells”). B cells contained fewer than 1% CD4+ or CD8+ cells. 8×104B cells were dispensed in triplicate into 96 well microtiter plates in 100 μl RPMI containing 10% FBS (heat inactivated to 65° C. for 30 min.), 50 μM 2-mercaptoethanol, 100 U / ml penicillin, 100 ug / ml streptomycin, and 2 mM L-glutamate. 20 μM ODN were added at the start of culture for 20 h at 37° C., cells pulsed with 1 μCi of 3H uridine, and harvested and counted 4 hr later. Ig secreting B cells were enumerated using the ALISA spot assay after culture of whole spleen cells with ODN at 20 μM for 48 h...
example 2
Effects of ODN on Production of IgM from B Cells
[0153] Single cell suspensions form the spleens of freshly killed mice were treated with anti-Thy1, anti-CD4, and anti-CD8 and complement by the method of Leibson et al., J. Exp. Med 154:1681 (1981)). Resting B cells (J. Exp. Med 155:1523 (1982). These were cultured as described above in 30 μg / ml LPS for 48 hr. The number of B cells actively secreting IgM was maximal at this time point, as determined by ELIspot assay (Klinman, D. M. et al. J. Immunol 144:506 (1990)). In that assay, B cells were incubated for 6 hrs on anti-Ig coated microtiter plates. The Ig they produced (>99% IgM) was detected using phosphatase-labeled anti-Ig (Southern Biotechnology Associated, Birmingham, Ala.). The antibodies produced by individual B cells were visualized by addition of BCIP (Sigma Chemical Co., St. Louis Mo.) which forms an insoluble blue precipitate in the presence of phosphatase. The dilution of cells producing 20-40 spots / well was used to dete...
example 3
B Cell Stimulation by Bacterial DNA
[0154] DBA / 2 B cells were cultured with no DNA or 50 μg / ml of a (Micrococcus lysodeikticus; b) NZB / N mouse spleen; and c) NSF / N mouse spleen genomic DNAs for 48 hours, then pulsed with 3H thymidine for 4 hours prior to cell harvest. Duplicate DNA samples were digested with DNASE I for 30 minutes at 37° C. prior to addition to cell cultures. E coli DNA also induced an 8.8 fold increase in the number of IgM secreting B cells by 48 hours using the ELISAspot assay.
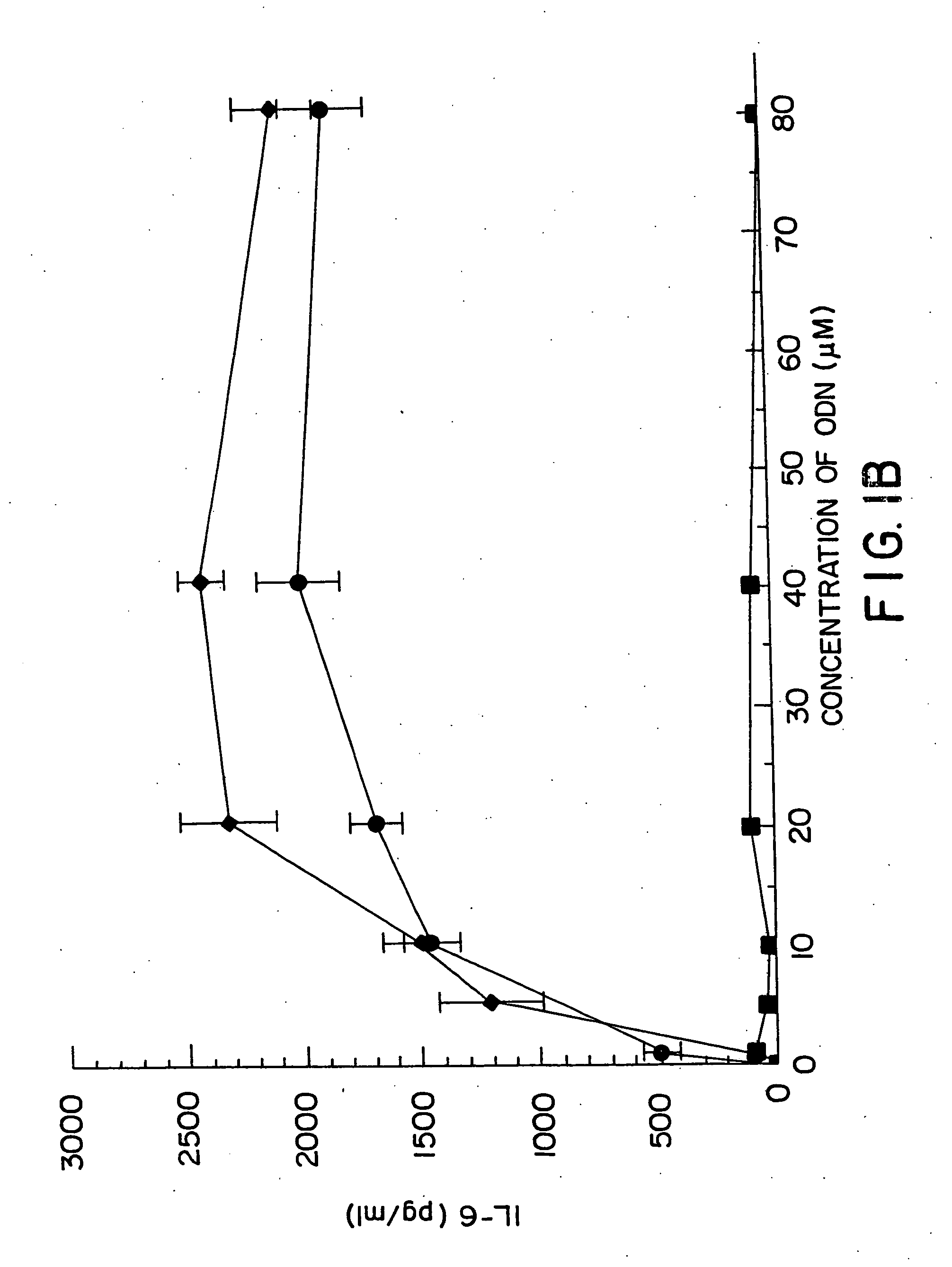
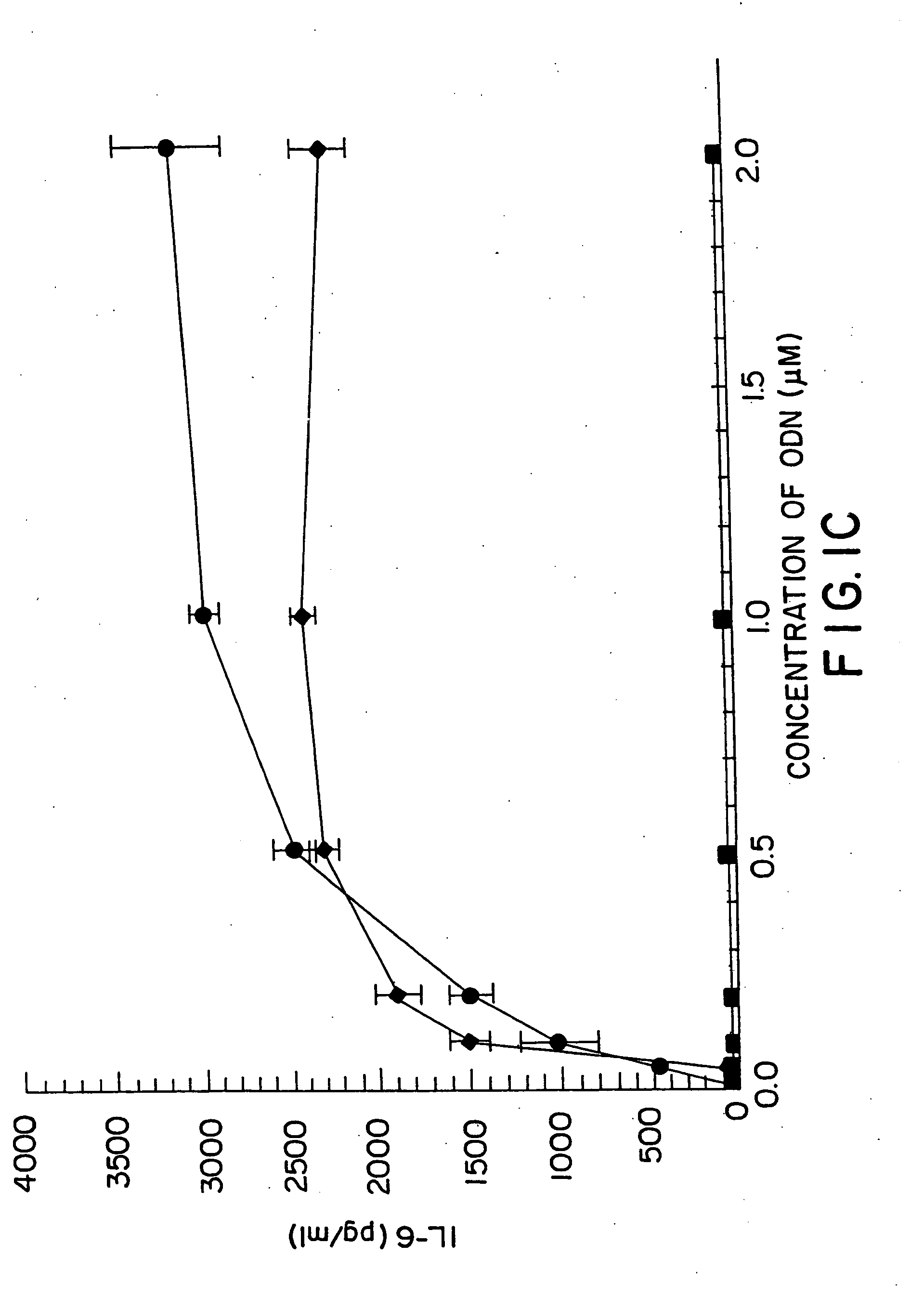
[0155] DBA / 2 B cells were cultured with either no additive, 50 μg / ml LPS or the ODN 1; 1a; 4; or 4a at 20 uM. Cells were cultured and harvested at 4, 8, 24 and 48 hours. BXSB cells were cultured as in Example 1 with 5, 10, 20, 40 or 80 μM of ODN 1; 1a; 4; or 4a or LPS. In this experiment, wells with no ODN had 3833 cpm. Each experiment was performed at least three times with similar results. Standard deviations of the triplicate wells were <5%.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| electron microscopy | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com