Target-specific multiple sequencing primer pool for microbial typing and sequencing applications in DNA-sequencing technologies
a technology of dnasequencing technology and primer pool, which is applied in the direction of microorganism testing/measurement, sugar derivatives, biochemistry apparatus and processes, etc., can solve the problems of difficult or impossible base calling, and achieve good results and improve sequence data quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials for HPV Typing
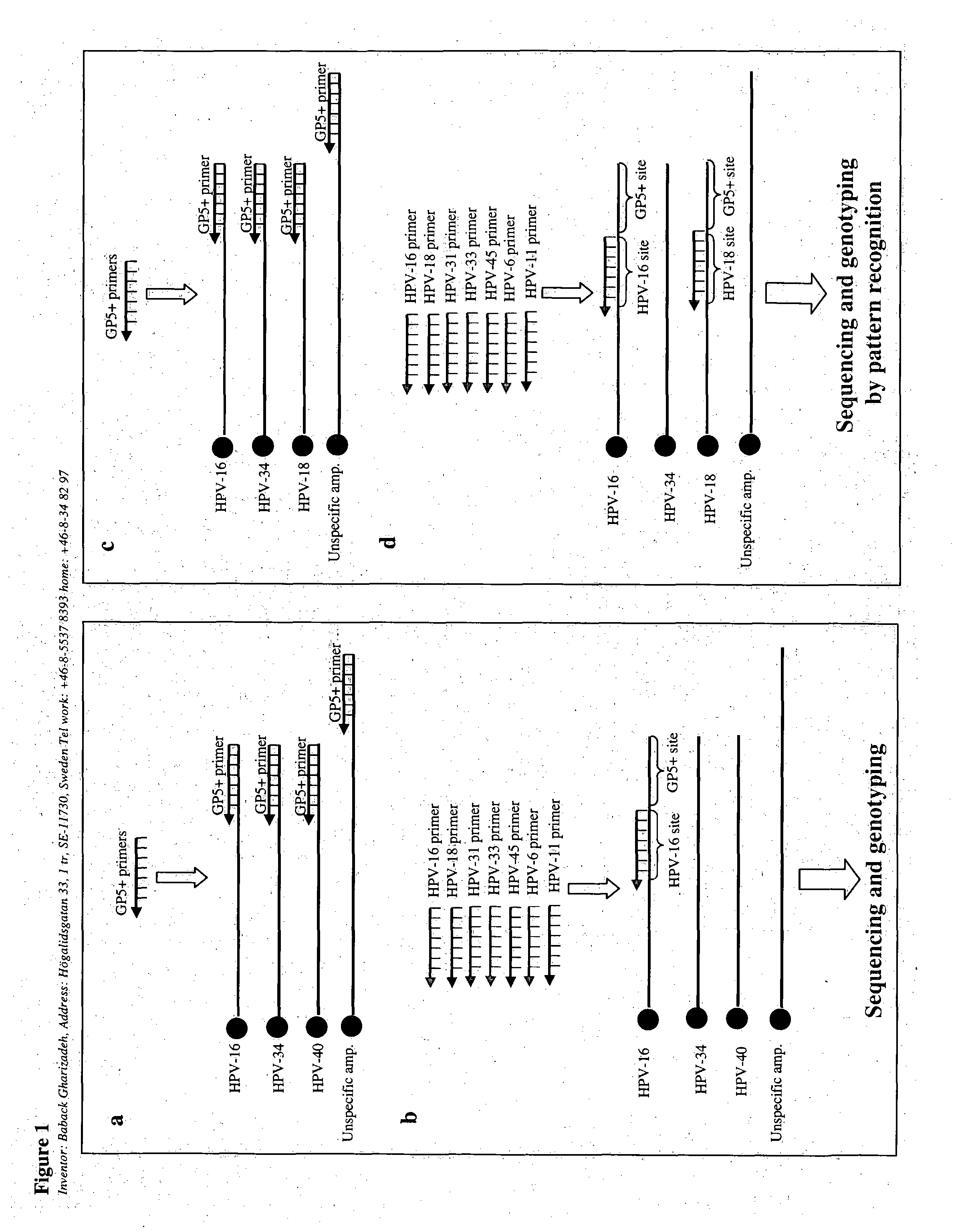
[0085] The primers for example 1 are designed for the HPV types that can be amplified with GP5+ / 6+ or MY09 / 11 primers sets. Each sequencing primer is designed to be specific for one designated HPV genotype. All the primers were checked by BLAST search for specificity and the resulting sequences were specific for the type designed.
[0086] The specific HPV-primers designed for detection of high-risk HPV by the multiple sequencing primer strategy of the invention were:
[0087] High-risk HPV genotypes
HPV-165′-GCTGCCATATCTACTTCAGAHPV-185′-GCTTCTACACAGTCTCCTGTHPV-315′-GTG CTG CAA TTG CAA ACA GTHPV-335′-ACACAAGTAACTAGTGACAGHPV-455′-TATGTGCCTCTACACAAAAT
[0088] Low-risk HPV genotypes
HPV-65′-GTG CAT CCG TAA CTA CAT CTTHPV-115′-GTG CAT CTG TGT CTA AAT CTG
[0089] The GP5+ / 6+ amplification primer set sequence is:
GP5+5′-TTT GTT ACT GTG GTA GAT ACT AC 3′Biotin-GP6+5′-GAA AAA TAA ACT GTA AAT CAT ATT C 3′
[0090] The MY09 / 11 amplification primer set sequence is:
Biotin-MY0...
example 2
Typing of HPV in Clinical Samples and Simulated Mixed-infections by Applying Multiple Sequencing Primers in Pyrosequencing Technology
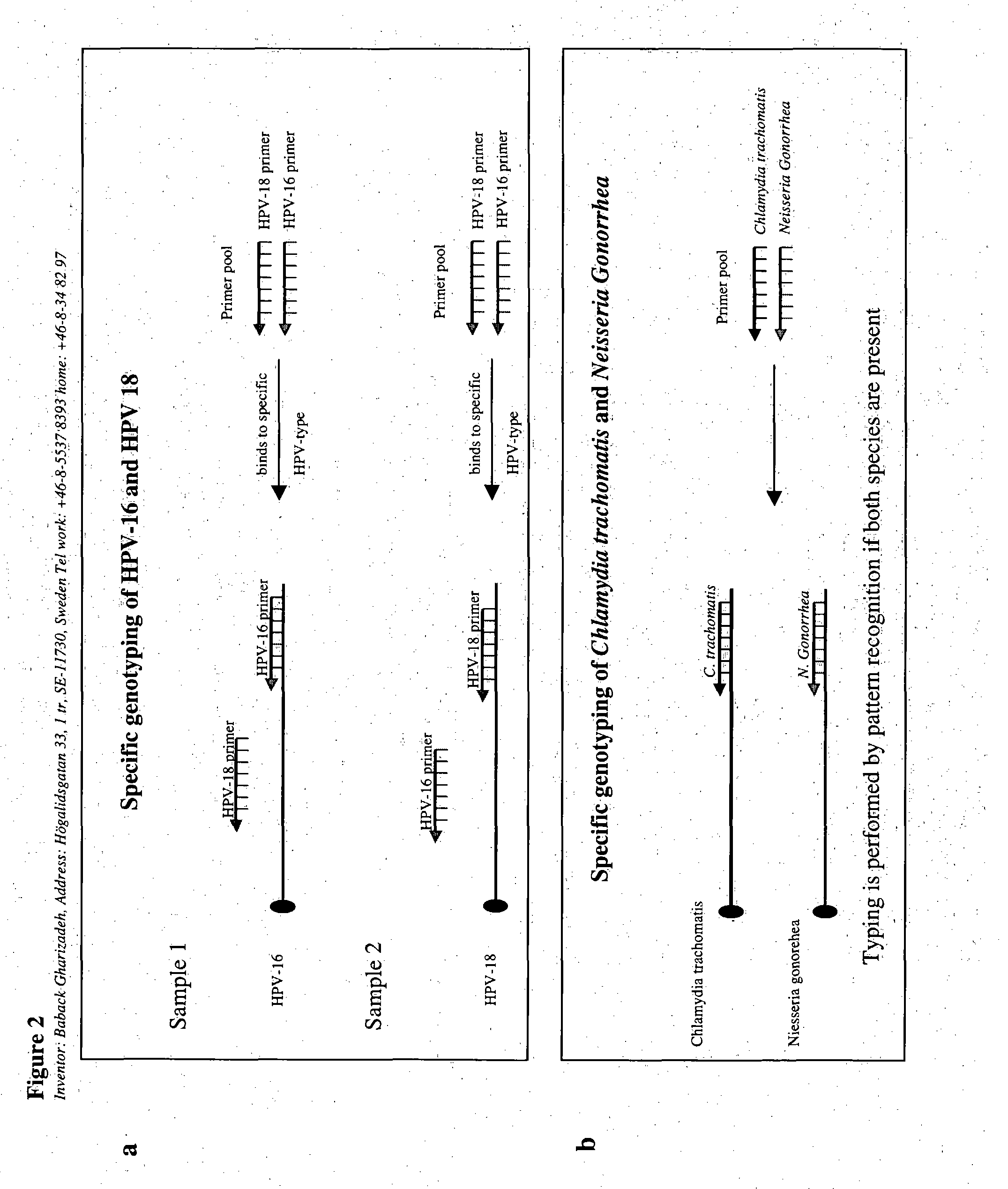
[0091] Some types of human papillomaviruses (HPV) are broadly recognized on a global scale as causative agents for cervical cancer. They are also related to other cancer types. For genotyping of HPVs we introduced the Pyrosequencing technology for specific HPV-typing (Gharizadeh et al., 2001). In general, not more than 25 bases are needed for specific genotyping. Multiple infections and unspecific amplification have been a problem for typing as double / multiple infections or unspecific amplification products present in one specimen give rise to sequence signals from all available types / products in the specimen. This is mainly because all HPV types in a sample are amplified with PCR utilizing the general GP5+ / 6+ primer set and then sequenced utilizing the GP5+ primer as extension / sequencing primer. This primer hybridizes to all present types, or unspeci...
example 3
Typing of HPV in Mixed-infections by Dideoxi DNA-sequencing (Sanger)
[0097] The general approach of example 2 was repeated, and Sanger sequencing was used as the DNA-sequencing method, instead of Pyrsosequencing (all the figures are not shown). FIG. 11a shows HPV-16 / 72 / 6-mixture sequenced with a general primer as extension primer. FIG. 11b shows HPV-16 / 72 / 6-mixture, sequenced by the primer set of the invention, which comprises a mix of 4 primers (HPV-16, 18, 33 and 45) (Gharizadeh et al., 2003). The results show clearly that with the dideoxy sequencing, typing with the primer set of the invention renders sequencing results that are clear to interpret than by using a general primer (impossible). An ABI prism DNA analyzer 3700 (Applied biosystems) was used for the DNA-sequencing with Big dye terminator kit according the manufacturer's manual. Same results were achieved on amplicons derived by MY09 / 11 using a-seven-multiple sequencing primers (unpublished data).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Distance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com