Simultaneous assay of target and target-drug binding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Antibody Optimization
[0034] The following method was employed to validate the method of the invention using the targets and antibodies described in Table 2, below. However, it can be applied to any novel combinations of antibodies to find suitable antibody pairs useful for any target-drug combinations of interest.
[0035] Commercially available conjugated antibodies against various antigens (generically called AgX) were identified and ordered for evaluation in the method of the invention. Non-conjugated, purified antibodies as well as proprietary pharmaceutical grade antibodies can also be obtained for evaluation and potential custom conjugation.
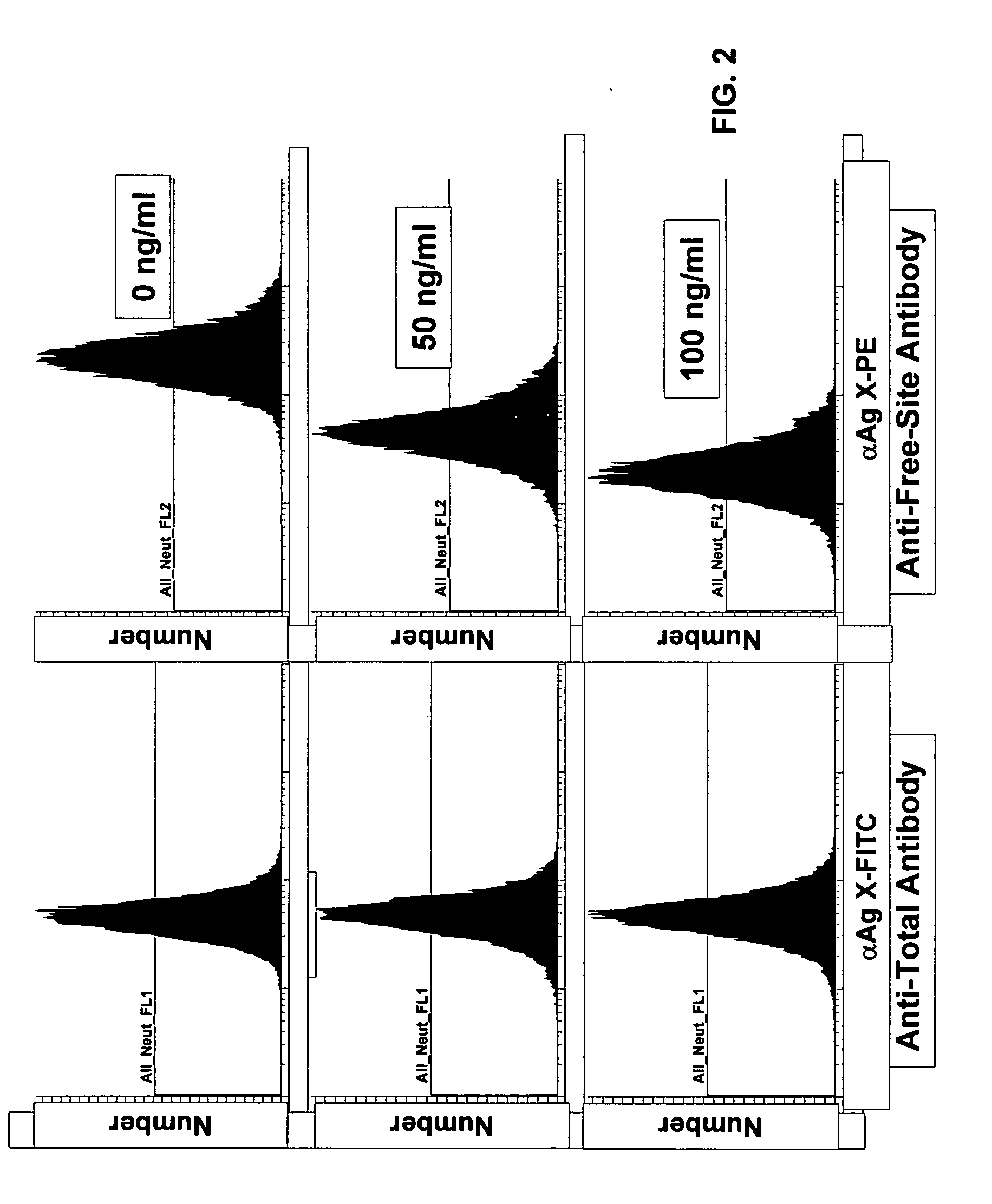
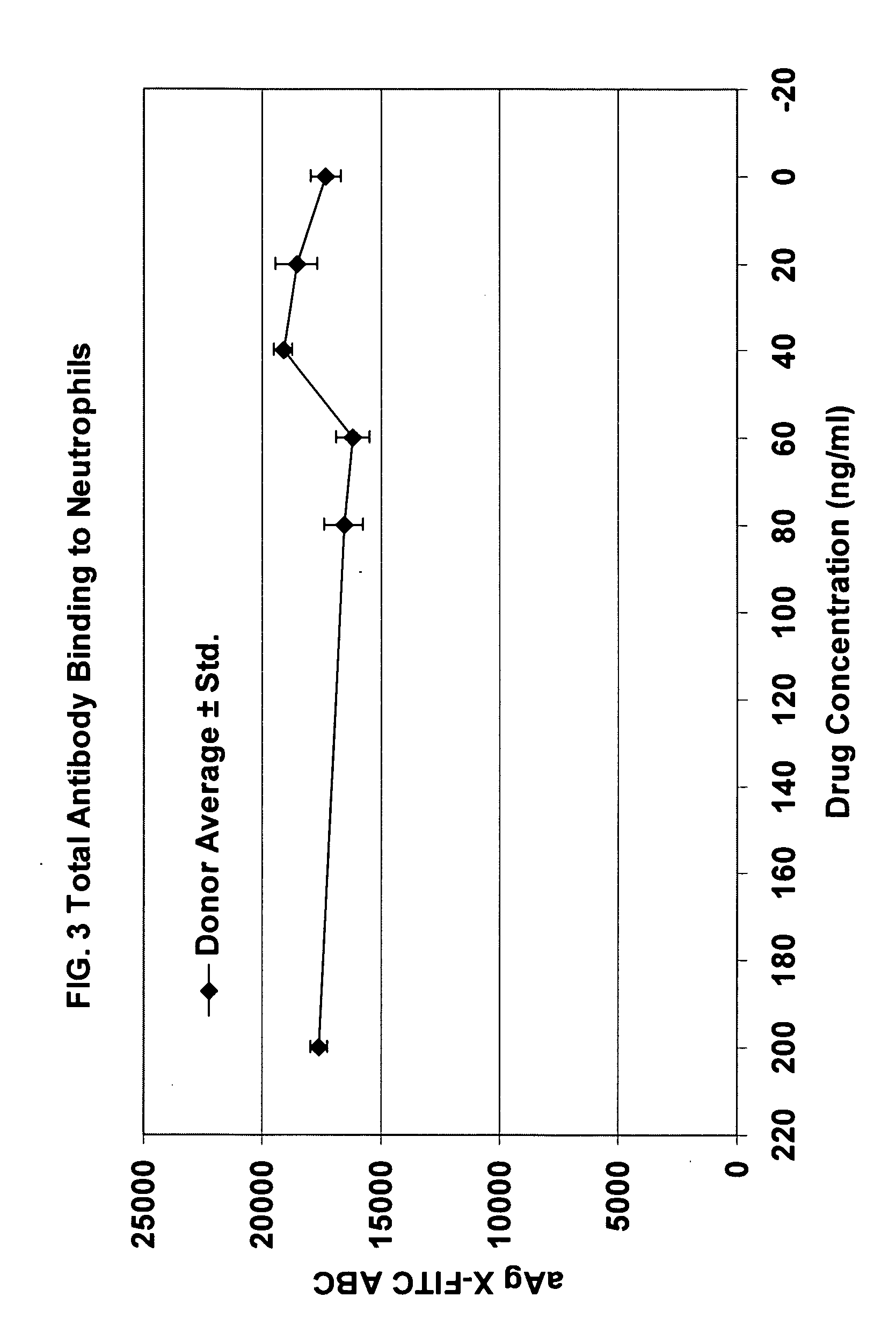
[0036] All anti-AgX antibodies against were evaluated in triplicate in the presence and absence of saturating concentrations of the appropriate drug (in each instance, the drug is specific for a single epitope on the protein AgX) by performing individual multi-point two-fold serial dilutions of each antibody. Additional cell subtype specifi...
example 2
Additional Antibody Pairs
[0041] Additional antibody pairs that can be used in the general method of the invention for particular target and drug combinations can be identified using the method described generally in Example 1. Some possible combinations are listed in Table 3. However, the combinations are unlimited and additional combinations can identified by searching MEDLINE, ATCC or the web.
TABLE 3Additional Target-Drug Combinations and Antibody PairsTargetDrugAntibodiesCD20Rituximab,PRO70769, Rituximab, BexxarBexxar(Tositumomab), B1, Immu-106,Ibritumomab, HI47, L27CD52Campath-1HCampath-1H(Alemtuzumab)CD33In DevelopmentHB-10306, Mab 251, M195,huM195, PC251, L4F3CD4In DevelopmentOKT4, TNX-355, HuMax-CD4,GK1.5, W3 / 25, YTS177.9,ORTHOCLONE OKTcdr4aCD3In DevelopmentHB231, HB10166, UCHT1, PS1,OKT3,CD14In DevelopmentHB-247, HB-246, UCHM-1, IC14,M5E2, MφP9CD30In DevelopmentcAC10-vcMMAE, SGN-30, Ki-1,K0145-3, HSR-4, BER-H2HLA-DrHu1D10Hu1D10, L243, BRA30, 1DO9C3,TNB-211, 1D10, LN3,MAP ...
example 3
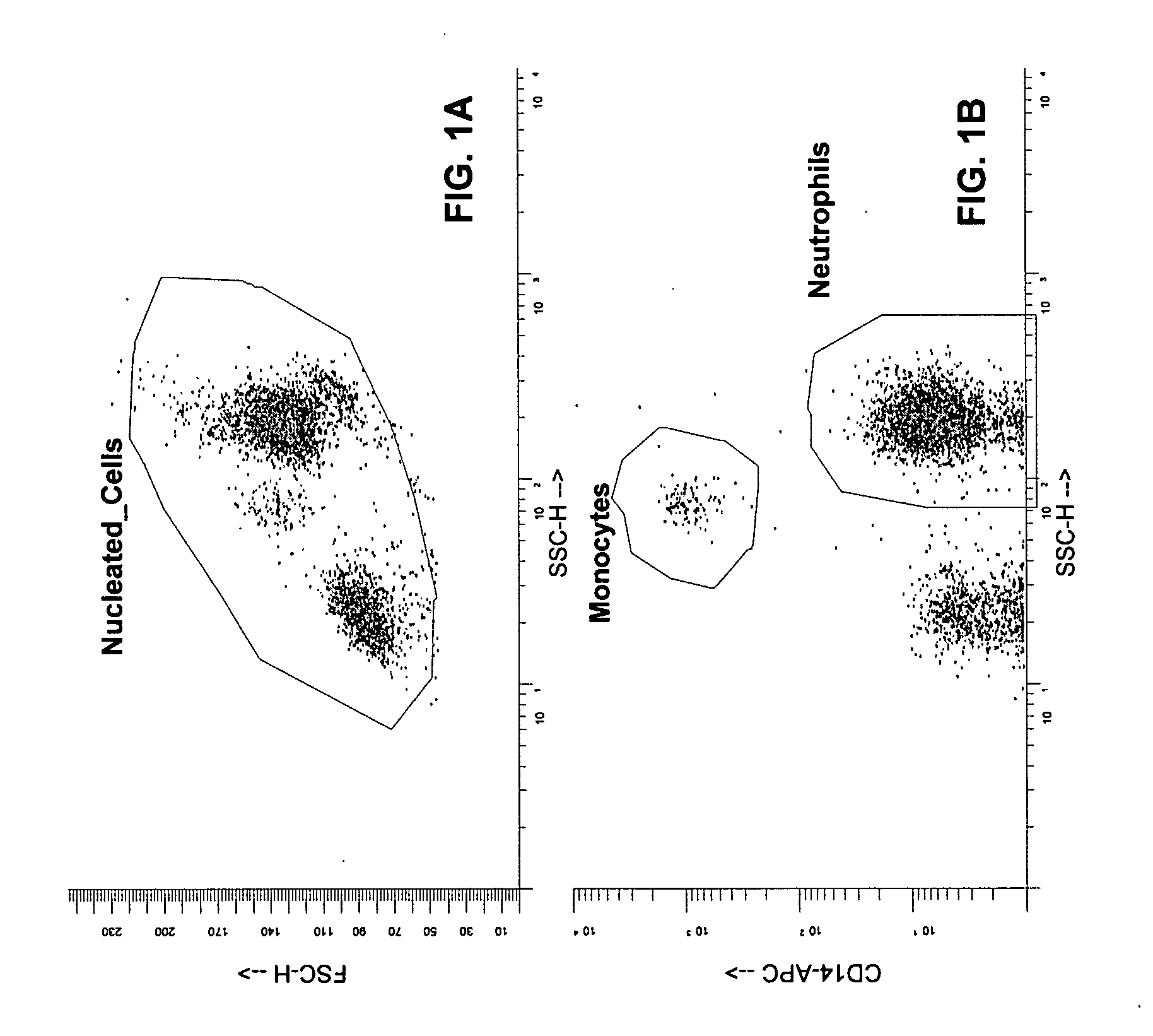
[0042] Although the invention has been exemplified with respect to several antibody pair-drug combinations, as indicated in Table 2, only the data from a single study (CD11b) are presented herein for simplicity. These results are representative, although the details for each study will vary. The protocols described are also exemplary, but cell harvesting, antibody staining, fixation, and gating parameters should be (and were) optimized for each experiment.
[0043] General Protocol: For analysis of whole blood by flow cytometry, the following protocol was employed: 100 μl of whole blood was sterilely collected for each data point. For each sample, 2 ml of standard culture media plus or minus drug was added to the blood and the tubes vortexed briefly to mix. The samples were then incubated to allow drug binding for 1 hr at 37° C.
[0044] Next the cells were stained with the appropriate antibody or antibodies. A saturating amount (as determined in the titration evaluation ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com