Stabilizing a nucleic acid for nucleic acid sequencing
a nucleic acid and nucleotide technology, applied in specific use bioreactors/fermenters, biomass after-treatment, biochemistry apparatus and processes, etc., can solve the problems of difficult implementation and inconvenient use of bulk sequencing techniques for the identification of subtle or rare nucleotide changes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Dual Biotinylation
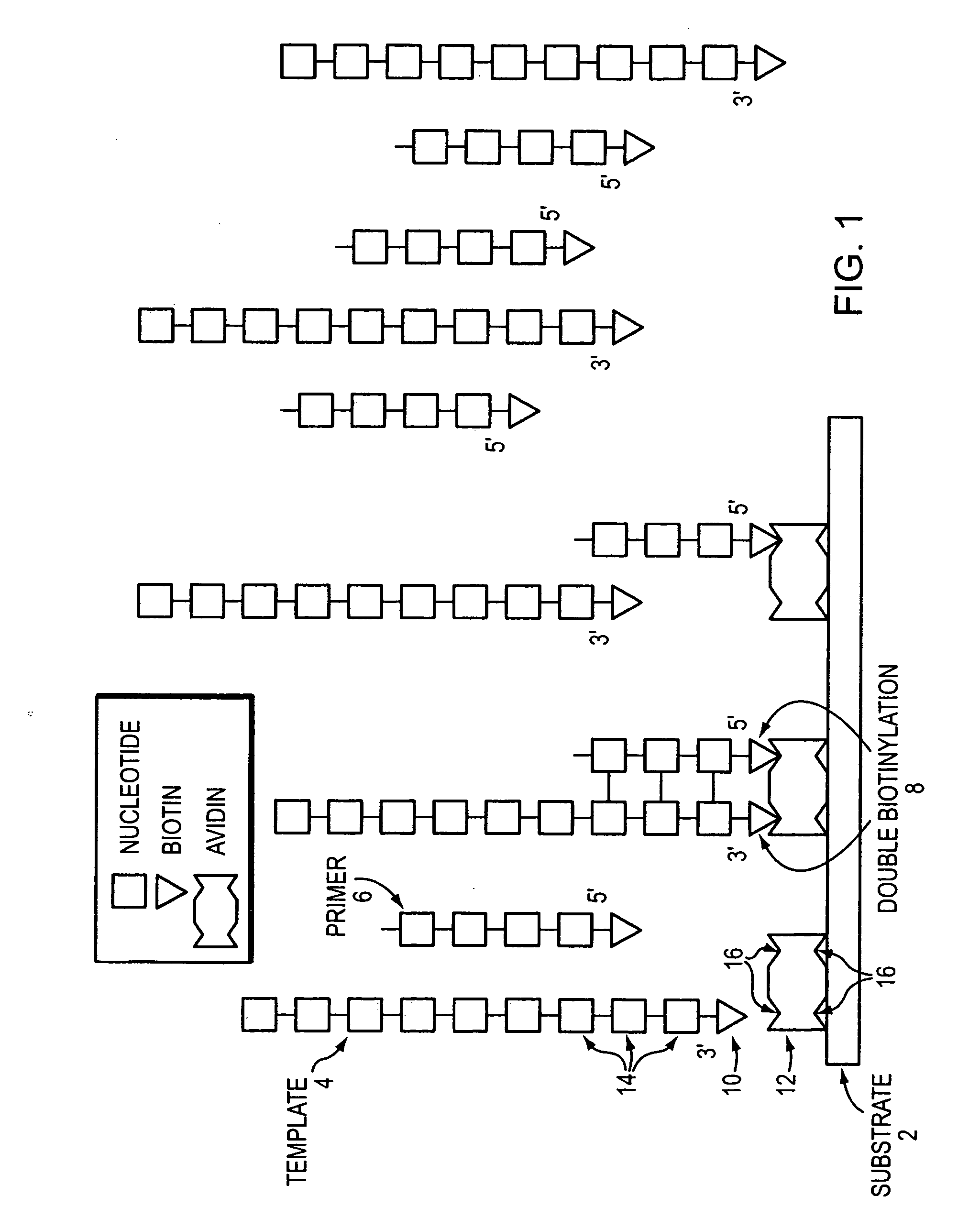
[0066] General methods of the invention were demonstrated using biotin / avidin binding pairs. When a biotin-streptavidin linkage is used to anchor a primer and a target nucleic acid to a substrate, the primer and target nucleic acid are biotinylated, while the surface of the substrate is coated with streptavidin. Because streptavidin is a tetramer, it is possible that both template and primer will bind to the same surface streptavidin. However, the dual biotin labels may bind to adjacent streptavidin molecules as well.
[0067] Two experiments were done to determine the binding stability of the dual biotin constructs. A first experiment was conducted in order to determine the stability of dual biotin duplex on a polyelectrolyte multilayer (PEM) surface. This experiment was done using covalent streptavidin attachment to a PEM surface. The PEM surfaces were prepared as follows. Polyethyleneimine (PEI) and pollyallylamine (PAA, Sigma, St. Louis, Mo.) were dissolved sepa...
example 2
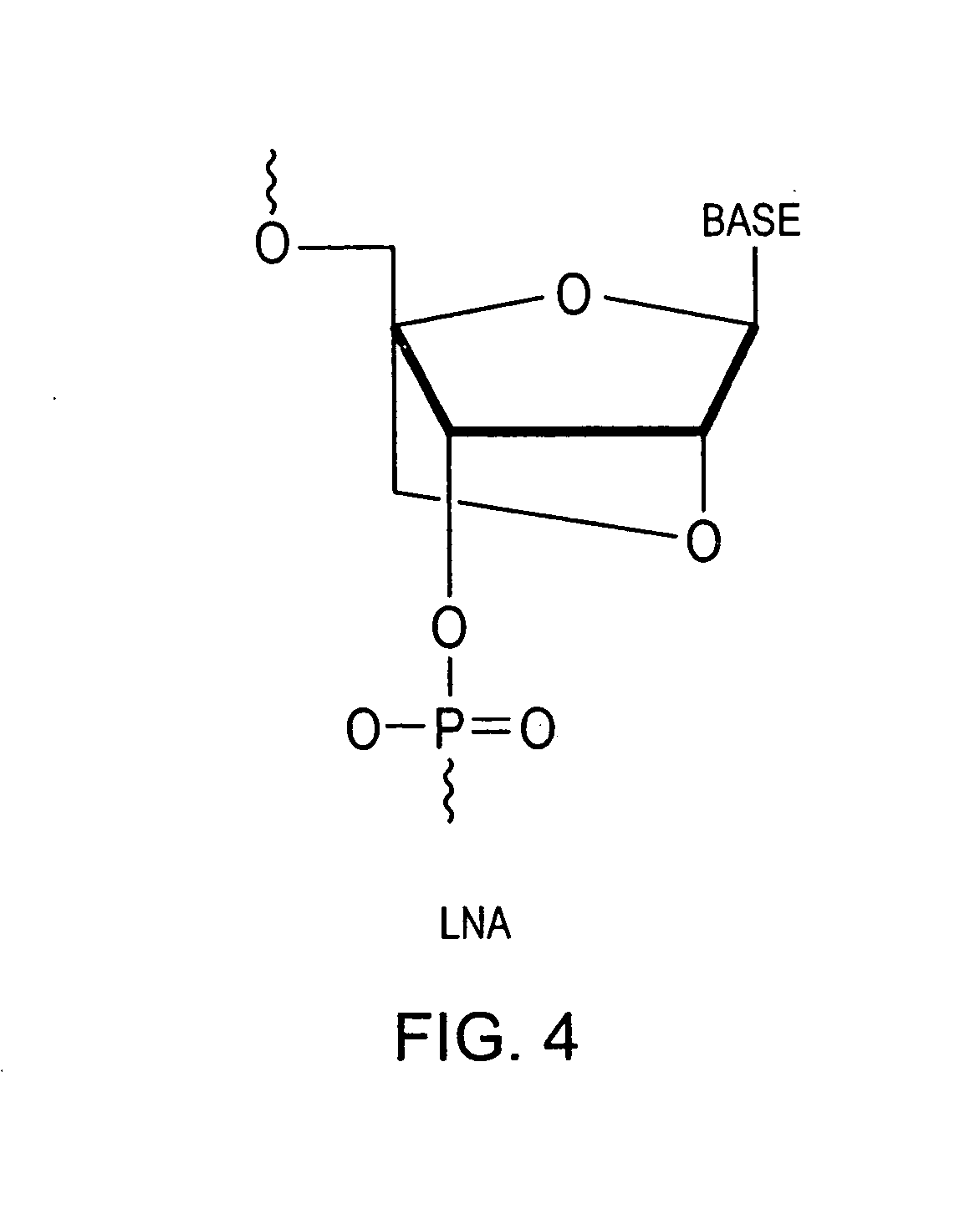
[0076] A primer is designed to be complementary to a known primer attachment site of the target nucleic acid, and locked nucleic acid bases are substituted for certain nucleotides within the selected primer sequence. As many locked nucleic acid bases are selected as desired depending on the temperature and length of primer, up to a primer comprising 100% locked nucleic acids. The more locked nucleic acid substitutions into the primer, the greater the melting point of the primer / target nucleic acid duplex relative a primer of the same length lacking locked nucleic acid residues.
[0077]FIG. 3 shows three primers synthesized with locked nucleic acids. DXS17, 7G7A, and 377 primers were synthesized as complementary strands to known regions of a template, each primer incorporating nine locked nucleic acids. When these primers are annealed to their respective templates, hybridization may be carried out at temperatures between about 80° C. to about 90° C.
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com