Oligonucleotide probes for detecting enterobacteriaceae and quinolone-resistant enterobacteriaceae
a technology of enterobacteriaceae and probes, applied in the field of diagnostic microbiology, can solve the problems of inability to establish i>providencia stuartii/i>, gyra mutations with fluoroquinolone resistance, and infection life-threatening,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
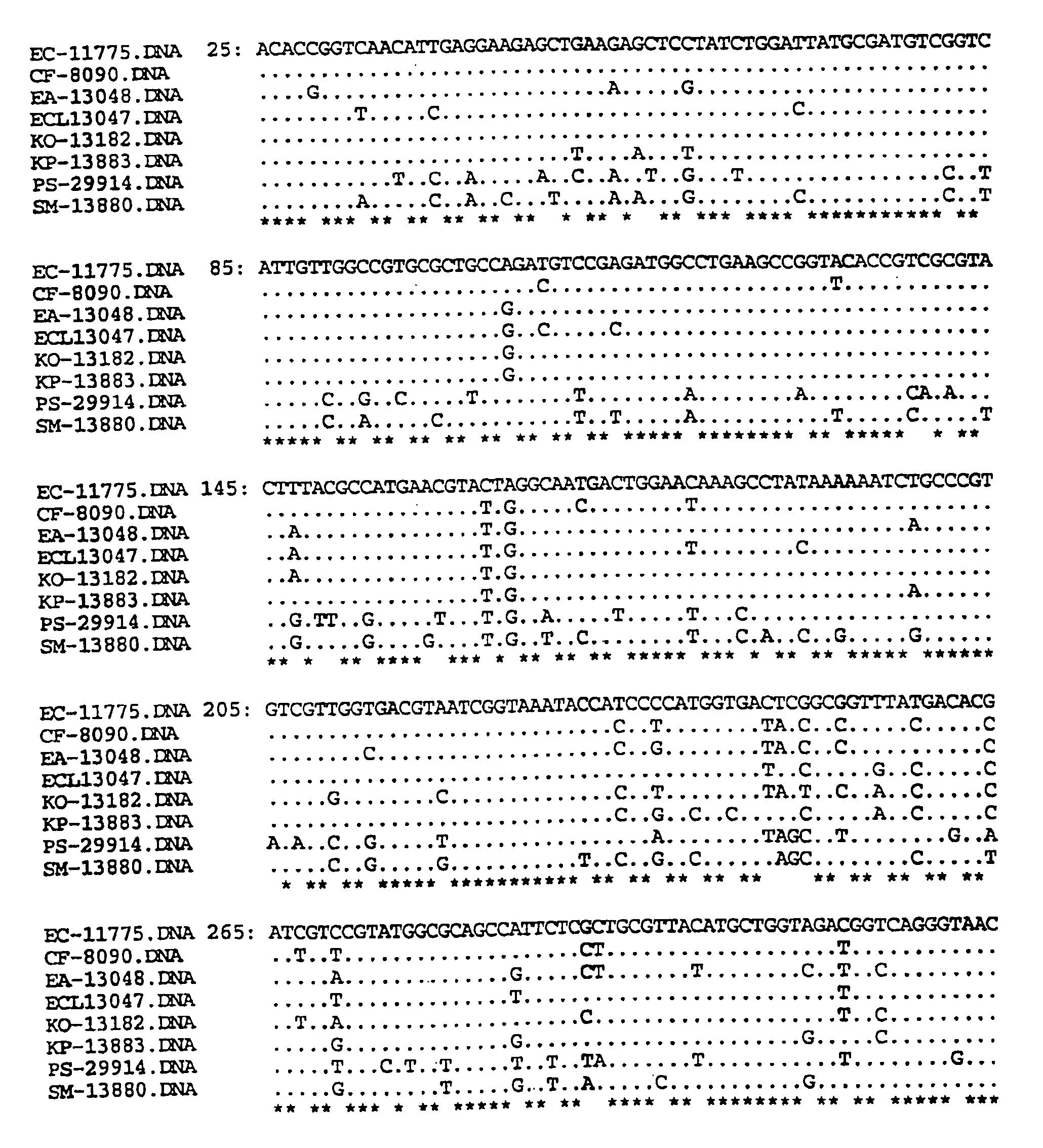
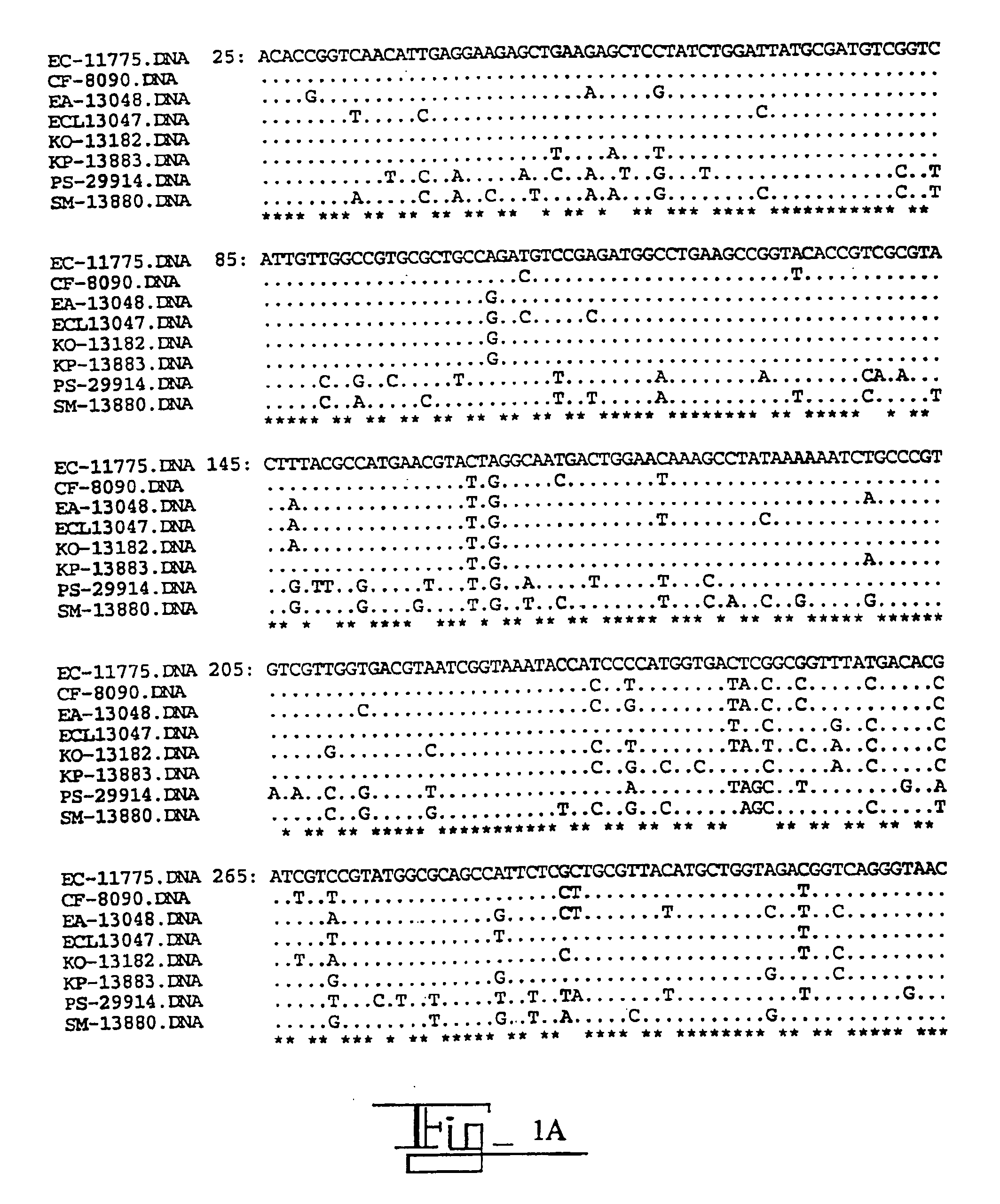
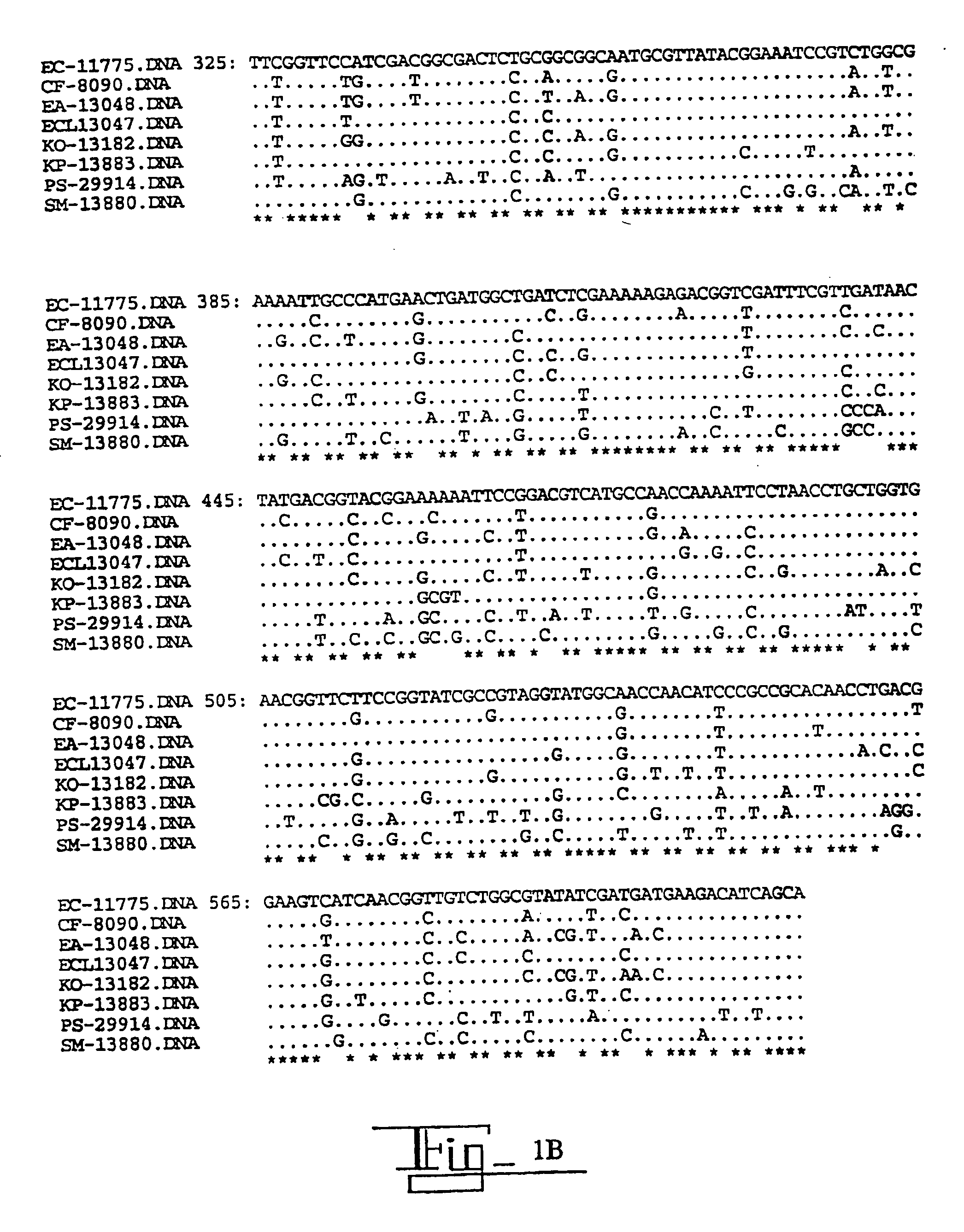
[0037]In this Example, the DNA sequence of the gyrA was determined for eight species of Enterobacteriaceae. Oligonucleotide primers were designed from conserved gyrA gene sequences flanking the QRDR and used to amplify and sequence the 5′ region of gyrA from ATCC type strains and fluoroquinolone-resistant clinical isolates. The nucleotide and the inferred amino acid sequences were aligned and compared.
[0038]The QRDR sequences from 60 clinical isolates with decreased fluoroquinolone susceptibilities were analyzed for alterations associated with fluoroquinolone resistance. The primer sequences at the 3′ and 5′ ends have been removed leaving nucleotides #25-613, based on the E. coli gyrA sequence numbers of Swanberg et al., J. Mol. Biol., 197:729-736 (1987). The organisms, abbreviations and ATCC type strain designation numbers are as follows.
EC=Escherichia coli (E. coli) ATCC 11775
CF=Citrobacter freundii (C. freundii) ATCC 8090
EA=Enterobacter aerogenes (E. aerogenes) ATCC 13048
ECL=Ente...
example 2
Development of Probes
Identification of Enterobacteriaceae Species
[0060]Oligonucleotide probes can be selected for species-specific identification of Enterobacteriaceae in or near the QRDR of gyrA. The region which includes the codons most often associated with fluoroquinolone resistance (nucleotides 239-263) was not used for the reason that if identification were based on one or more nucleotide changes, the changes associated with resistance would interfere with identification. Each probe for identification was selected for maximum difference, and it is recognized that a smaller region within some probes could be used, based on single base changes. However, most of the probes have at least two nucleotide differences compared with the same region in other strains. When there were variations, other than those associated with resistance, within the susceptible and / or the resistance strains for any given species, the position of the probe was shifted to a region which was completely con...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com