Data analysis to provide a revised data set for use in peptide sequencing determination
a technology of peptide sequencing and data analysis, which is applied in chemical methods analysis, separation processes, instruments, etc., can solve the problems of ecd not being able to perform ecd with trap-type mass analyzers, their own problems, and expensive instruments, so as to improve the confidence in precursor identification, cost less, and less time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
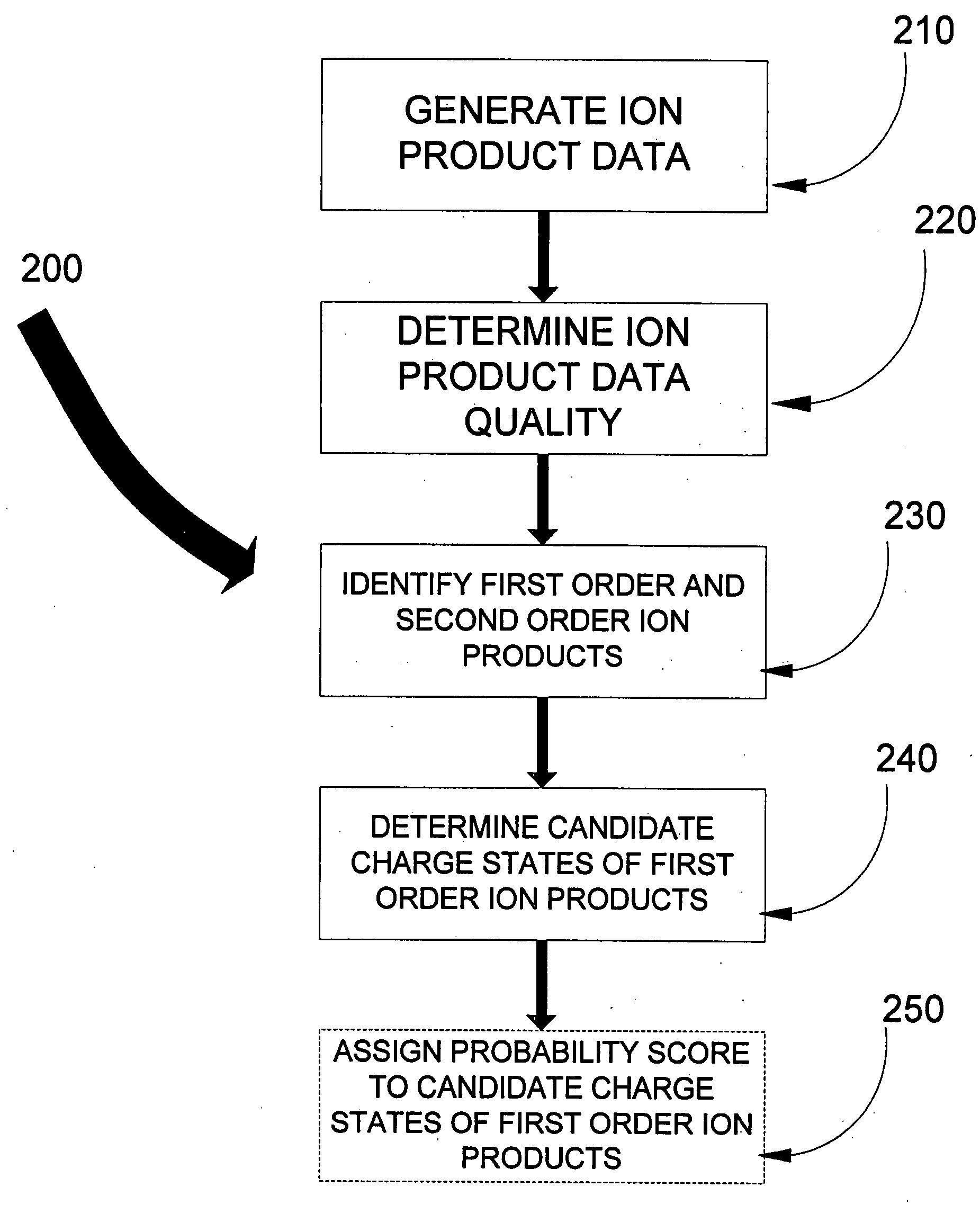
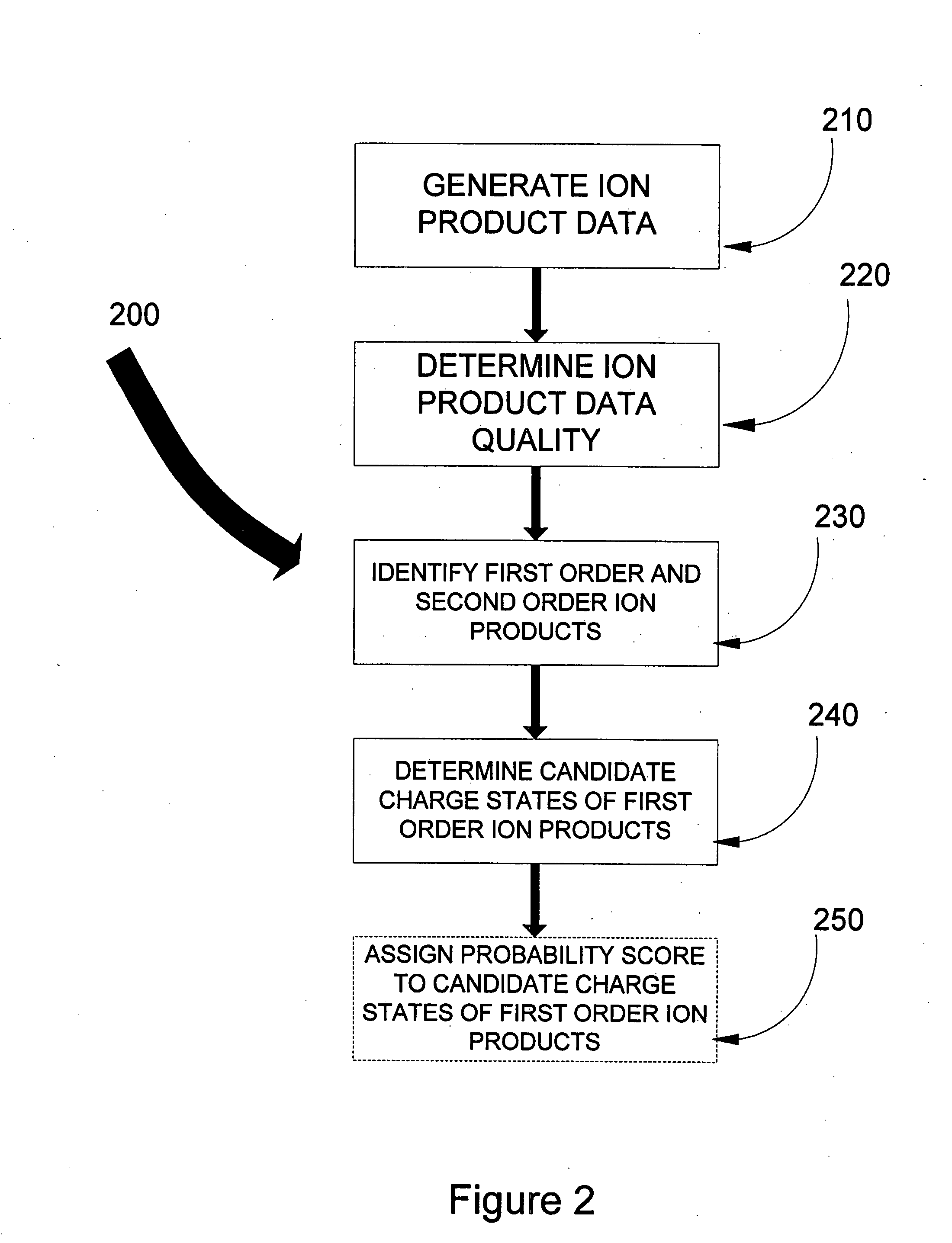
[0025]The present invention addresses some of the shortcomings of the known art. A method for determining the charge states of product ions generated from precursor ions by a non-ergodic technique such as ETD or ECD is provided in one aspect of the invention. In another, an improved method for determining the charge state of a precursor ion for use in peptide sequencing determination is also provided.
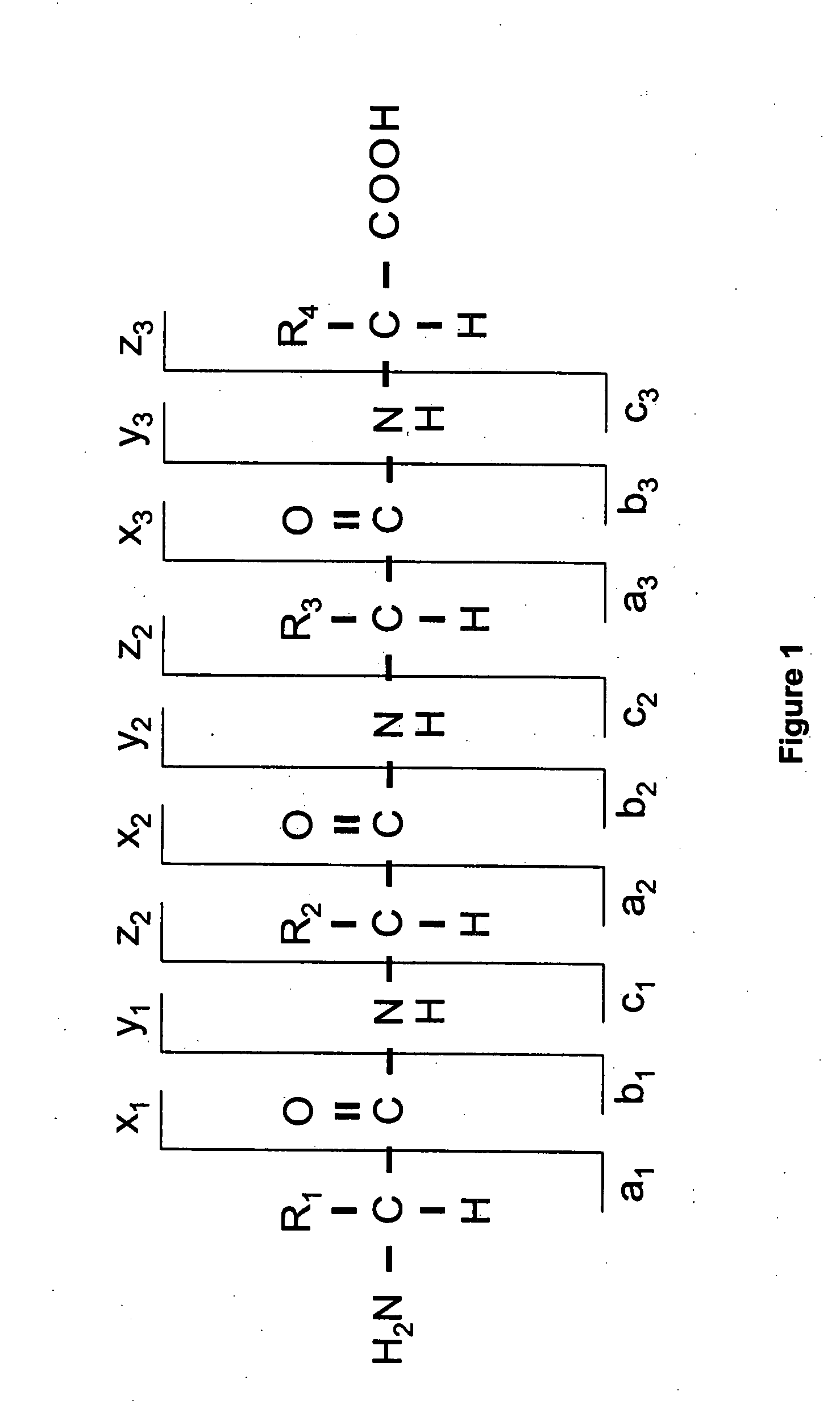
[0026]Before describing the invention in detail, a few terms that are used throughout the description are explained. As used in this specification, a peptide or polypeptide is a polymeric molecule containing two or more amino acids joined by peptide (amide) bonds. As used in this specification, a peptide typically represents a subunit of a parent polypeptide, such as a fragment produced by cleavage or fragmentation of the parent polypeptide using known techniques. Peptides and polypeptides can be naturally occurring (e.g., proteins or fragments thereof) or of synthetic nature. Polypepti...
PUM
| Property | Measurement | Unit |
|---|---|---|
| mass spectra | aaaaa | aaaaa |
| charge | aaaaa | aaaaa |
| mass-to-charge ratio | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com