Compositions and Methods of Plant Breeding Using High Density Marker Information
a marker information and high density technology, applied in knowledge representation, instruments, computing models, etc., can solve the problems of limiting inferences, reducing the efficiency of breeding programs, and reducing the efficiency of plant breeding, so as to improve the germplasm of plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
An Example of Haplotype-Trait Association Analysis: Grain Yield QTL on Chromosome 4 in Corn
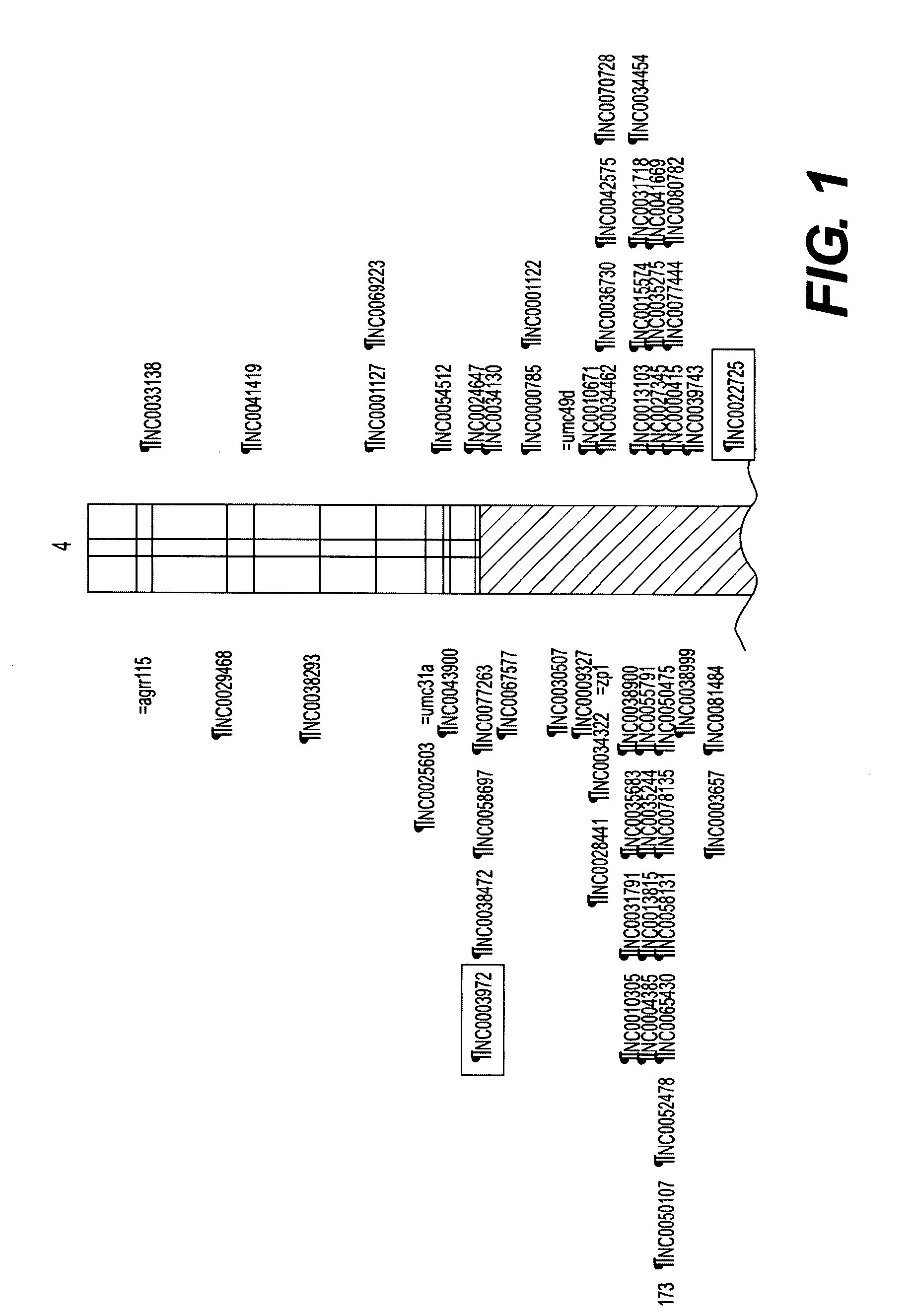
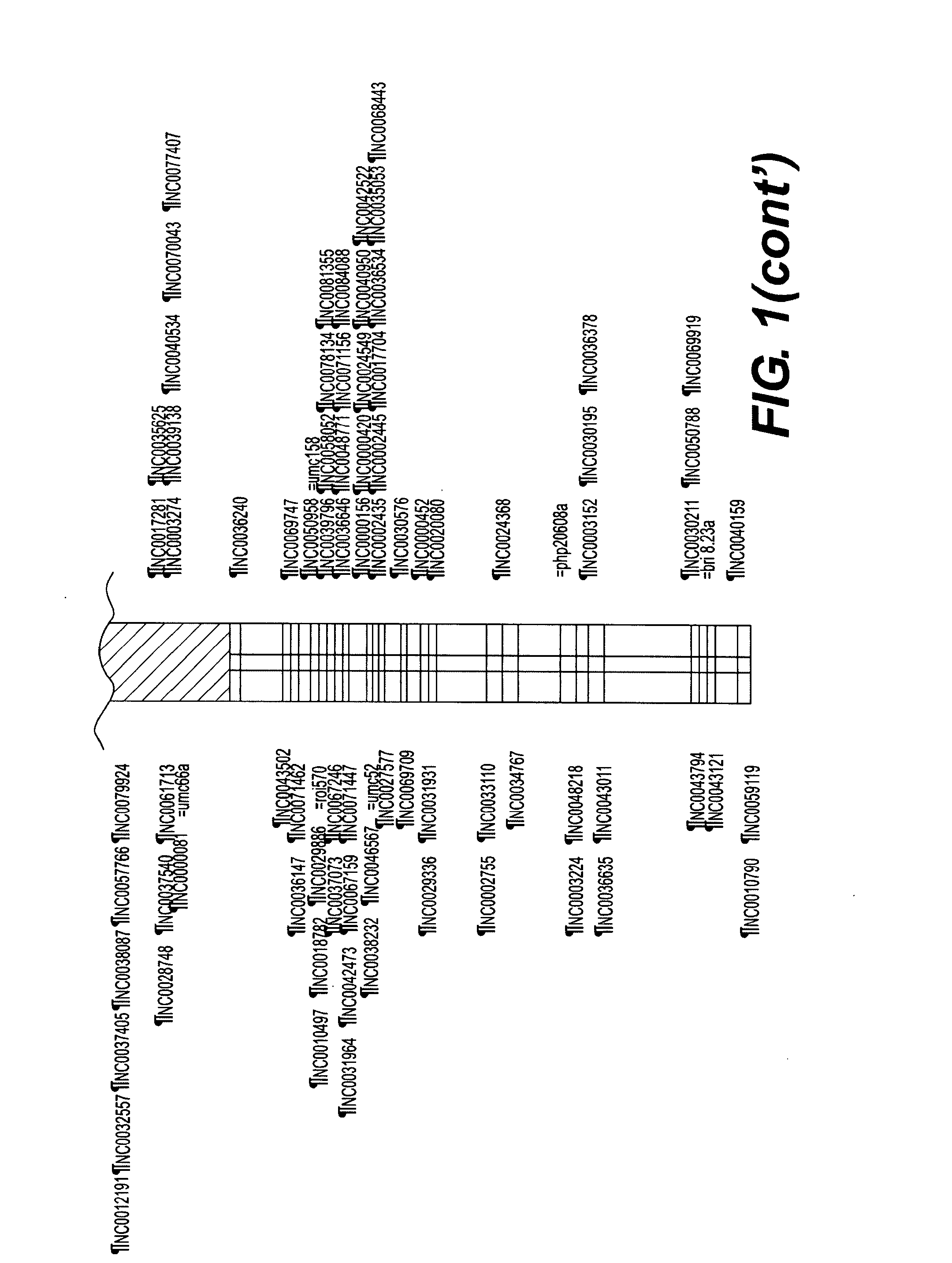
[0118]A key benefit of associating traits at the haplotype, rather than marker, level is the degree of resolution achieved. An initial QTL analysis from two different breeding crosses projects (herein denoted 1 and 2) were yield tested at 8 locations. A QTL was identified for grain yield on Chromosome 4 located approximately between 48 and 78 cM. The estimated QTL effect was similar in magnitude (4.2 Bu / Acre) for both projects. In the project 1, the genomic region from the inbred 5750 increased grain yield relative to the genomic region from the inbred 3140 when tested on the inbred 7051. In the project 2, the genomic region from the inbred 3323 increased grain yield relative to the genomic region from the inbred 90LDC2 when tested on the inbred WQDS7. The current breeding methodology uses this type of information (marker-QTL associations) to do recurrent selection within each population (proj...
example 2
Use of Breeding Values for Informing Decisions in a Breeding Program
[0121]A primary innovation of the present invention is the ability to simultaneously select for multiple traits and target regions throughout the genome. Furthermore, this invention leverages historical marker-phenotype information, enabling pre-selection.
[0122]A key aspect of predictive haplotype-assisted selection is the ability to rank haplotypes. This example includes a subset of 10 preferred haplotypes, across 10 haplotype windows, for yield from elite temperate female corn inbreds that have been ranked using haplotype breeding value calculations. The haplotype effect estimates for each of the haplotypes for six different phenotypic traits is shown in Table 10. This example illustrates the application of breeding values in decisions relating to germplasm improvement.
TABLE 10Haplotype effect estimates for six traits in 10 haplotypes from 10 differenthaplotype windows based on historical haplotype-phenotype assoc...
example 3
Implementation of Pre-Selection in a Breeding Program Via Automatic Model Picking
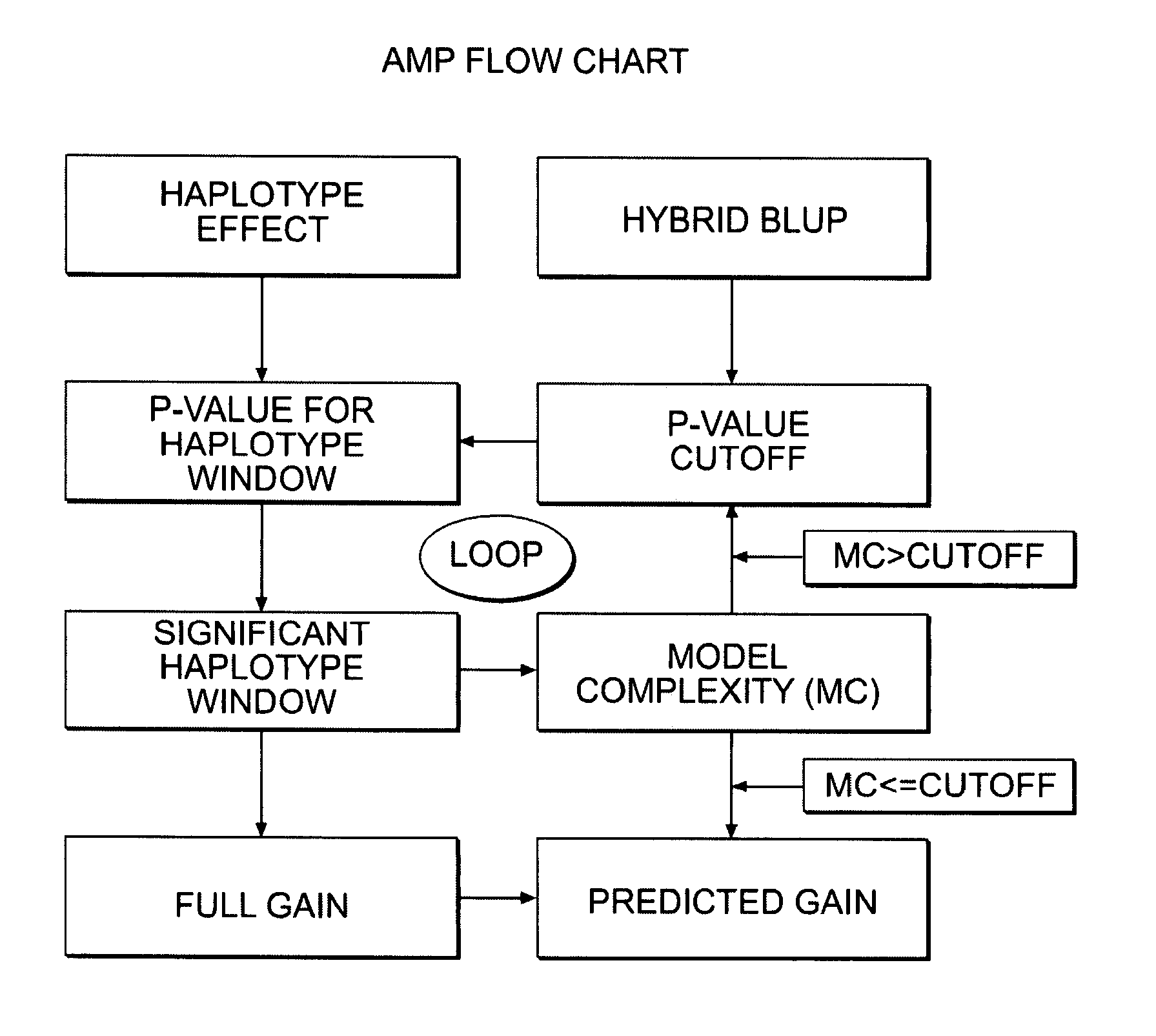
[0126]With haplotype estimation, pre-selection can be applied to a breeding program. This enables breeders, through marker-assisted selection on pre-determined significant haplotypes, to make genetic gain before new lines and breeding crosses are tested in the field. Breeders start pre-selection projects by selecting a list of crosses and building models based on the haplotypes carried by each parental line in the cross. One approach is to manually select haplotypes, but this hampers the breeders' ability to sort through a large number of possible crosses. There may also be inconsistencies in the way haplotypes are selected from cross to cross and there may be a need to restrain the choice of too many genomic regions in the model. For instance, if the model is too complex, predictive ability, and potential genetic gain, will likely be compromised. To control for model complexity and also meet high-throu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| disease resistance | aaaaa | aaaaa |
| pest resistance | aaaaa | aaaaa |
| stress tolerance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com