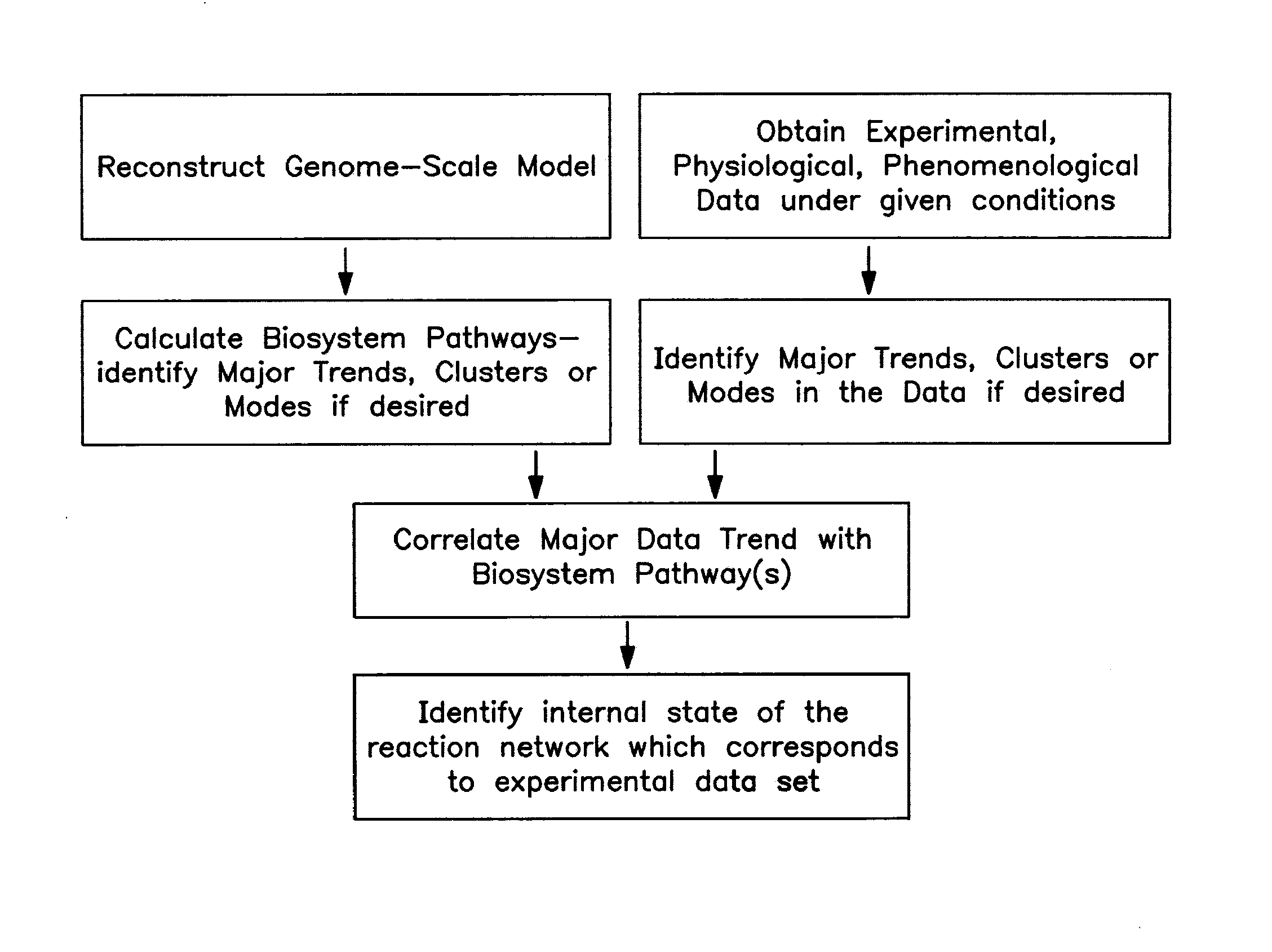
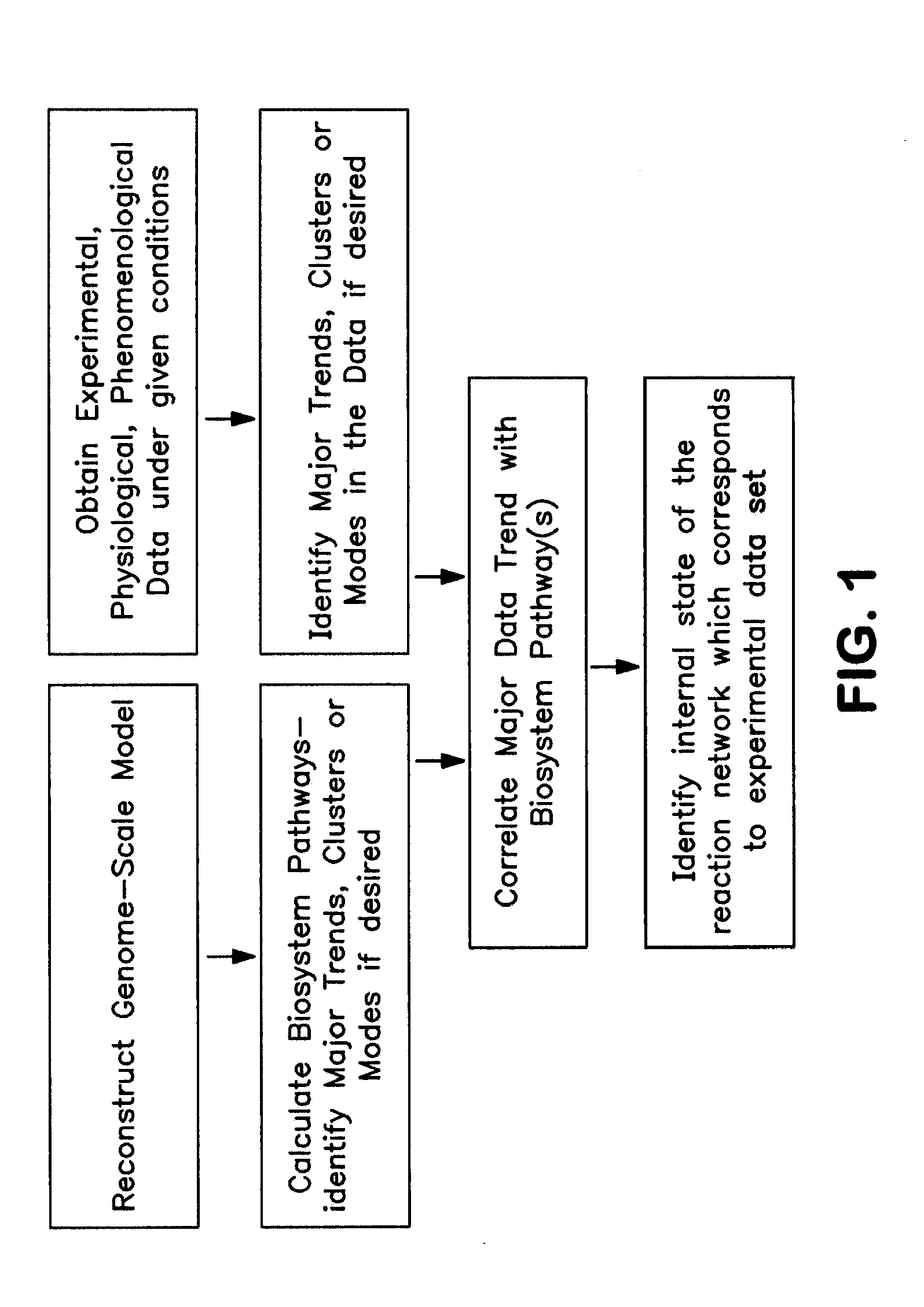
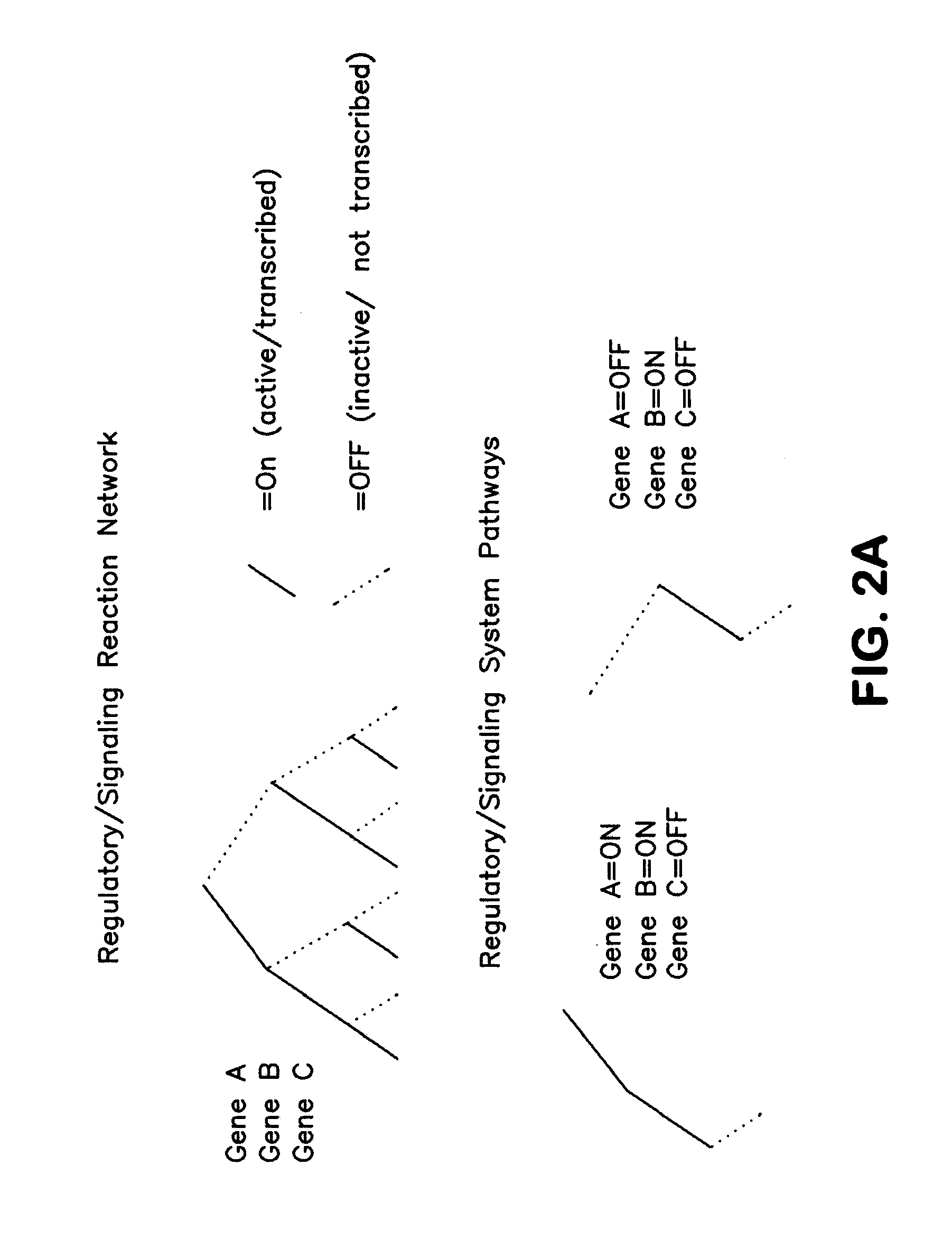
Methods and Systems to Identify Operational Reaction Pathways
a technology of operational reaction and method, applied in the field of in silico model organism construction, can solve the problems of difficult to predict a priori the effect of a single gene or gene product change, or the effect of a drug or an environmental factor, on cellular behavior, and current approaches to the development of compounds and processes used in these directions do not take into account the level of accuracy needed for perturbation effect on cellular behavior,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
Decomposing a Set of Phenomenological Flux Distributions for the E. coli Core Metabolic Network in Order to Identify Operational Extreme Pathways
[0140]This example shows how a set of phenomenological pathways (flux distributions) can be decomposed into dominant modes these modes can be compared with a set of systemic pathways (extreme pathways) to identify operational reaction pathways of a metabolic reaction network (E. coli core metabolism).
[0141]An in silico-generated metabolic flux profile of core metabolism in E. coli was prepared. The reactions were taken from table 6.3 of Schilling, “On Systems Biology and the Pathway Analysis of Metabolic Networks,” Department of Bioengineering, University of California, San Diego: La Jolla. p. 198-241 (2000), with the exception that reaction pntAB was not included, and instead of T3P2 in reaction tktA2, T3P1 was used. The reaction list is tabulated in Table 1.
[0142]The flux profile, which is the input matrix for Singular Value Decomposit...
example ii
Identifying Human Red Blood Cell Extreme Pathways Corresponding to Physiologically Relevant Flux Distributions
[0151]This example shows how a set of phenomenological pathways (flux distributions) generated by a kinetic model can be compared with the modal decomposition of a set of systemic pathways (extreme pathways) to identify dominant regulatory modes of a metabolic reaction network (human red blood cell metabolism).
[0152]The extreme pathways of the red blood cell (RBC) metabolic network have been computed (Wiback, S. J. & Palsson, B. O. Biophysical Journal 83, 808-818 (2002)). Here, SVD analysis was applied to the extreme pathway matrix, P, formed by these pathways. A full kinetic model of the entire metabolic network of the RBC has been developed (Jamshidi, N., Edwards, J. S., Fahland, T., Church, G. M., Palsson, B. O. Bioinformatics 17, 286-7 (2001); Joshi, A. & Palsson, B. O. Journal of Theoretical Biology 141, 515-28 (1991)), and was used to generate flux vectors (v) for ph...
example iii
In Silico Assessment of the Phenotypic Consequences of Red Blood Cell Single Nucleotide Polymorphisms
[0171]The following example illustrates the application of the described methods to analysis of phenomenological pathways defined through pathological data.
[0172]The Human Genome Project (HGP) is now essentially complete. One result of the HGP is the definition of single nucleotide polymorphisms (SNPs) and their effects on the development of human disease. Although the number of SNPs in the human genome is expected to be a few million, it is estimated that only 100,000 to 200,000 will effectively define a unique human genotype. A subset of these SNPs are believed to be “informative” with respect to human disease (Syvanen, A., 2001. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat Rev Genet 2: 930-942). Many of these SNPs will fall into coding regions while others will be found in regulatory regions. The human genotype-phenotype relationship is very comple...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
| chemical conversion | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com