Multiparameter FACS Assays to Detect Alterations in Cellular Parameters and to Screen Small Molecule Libraries
a technology of cellular parameters and facs assays, applied in the field of new methods of detecting alterations in cellular parameters and screening libraries of small molecules, can solve the problems of accelerating the accumulation of mutations driving malignant transformation, reducing the background of assays, and increasing specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
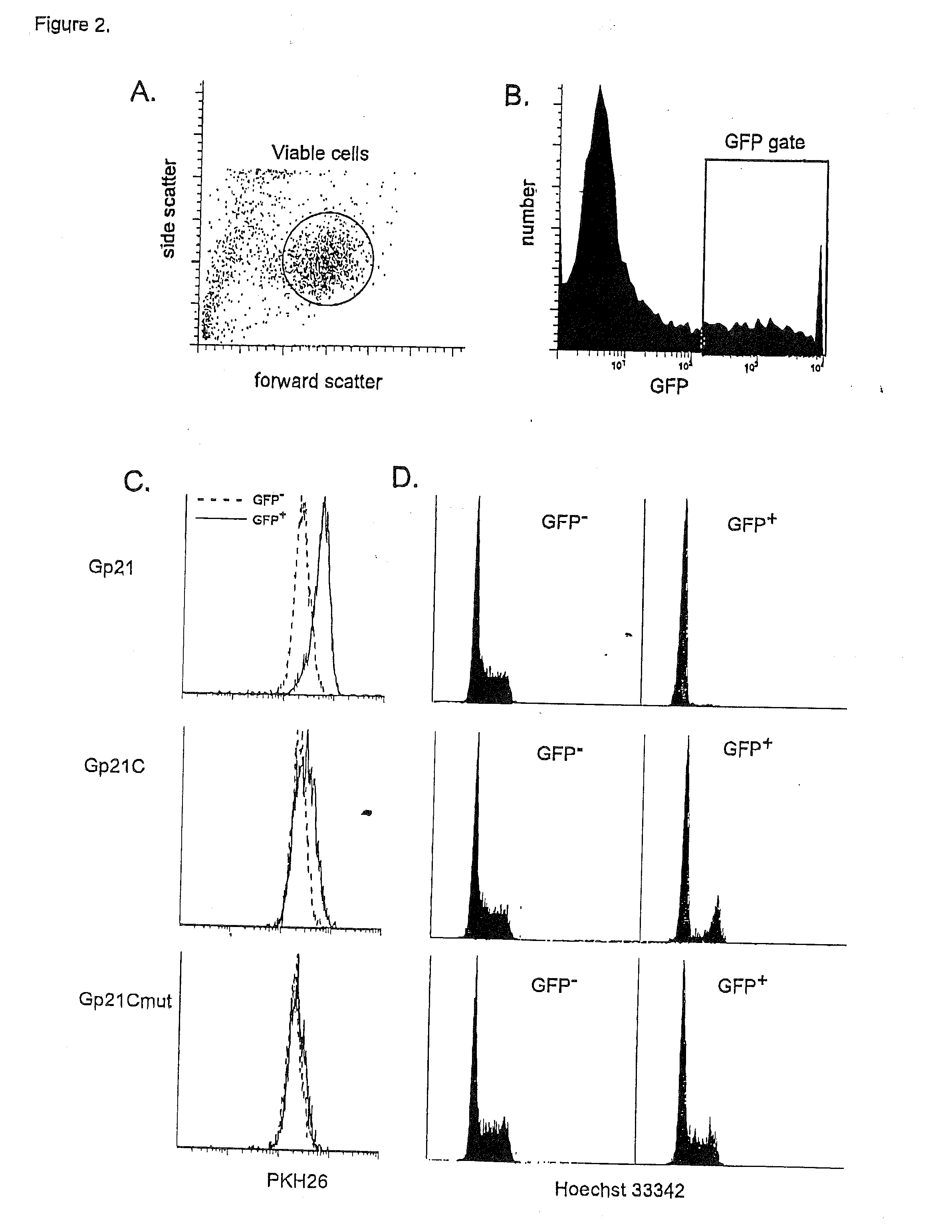
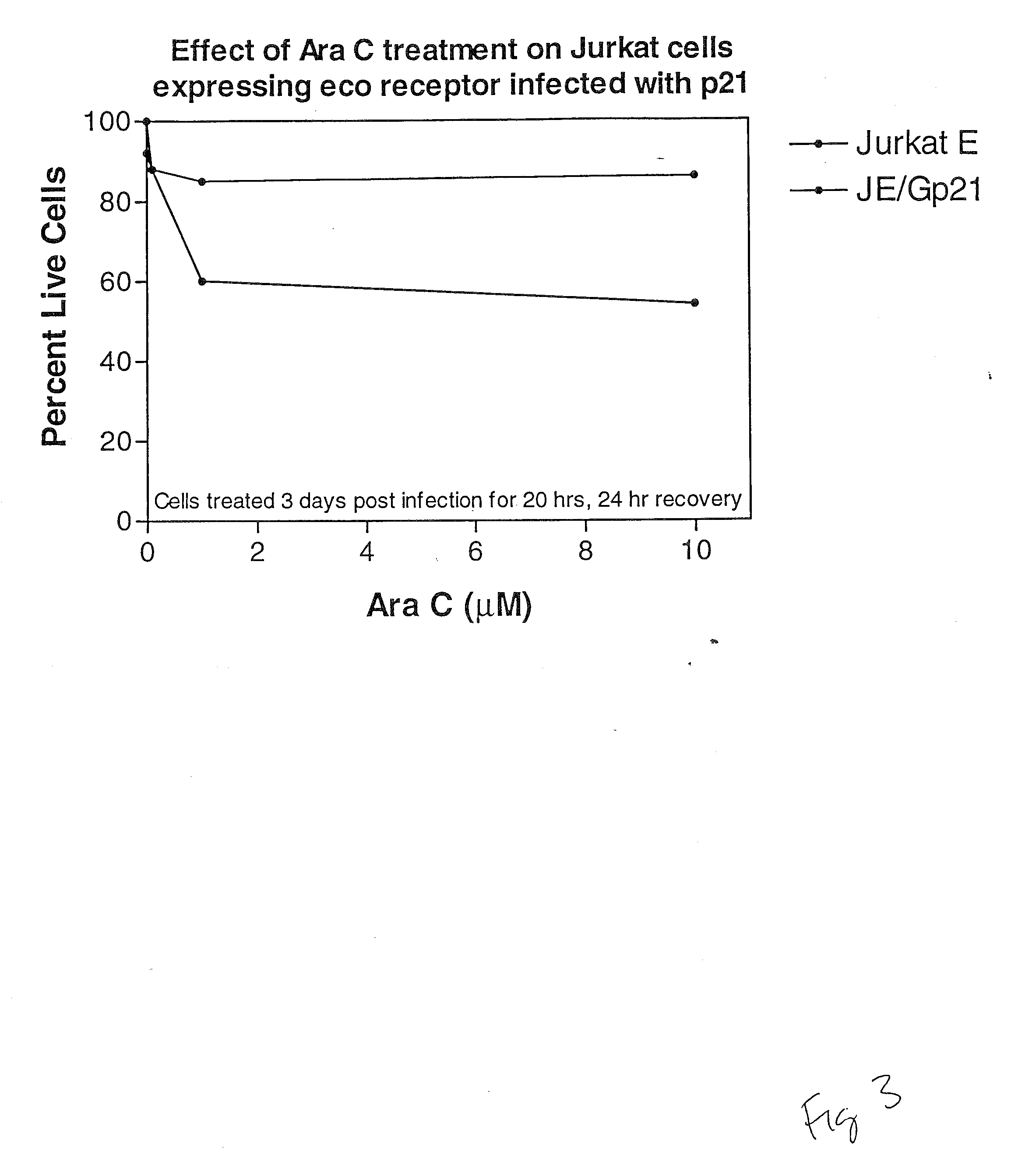
Cell Cycle Assays Using p21 as a Positive Control
[0194]Materials and Methods:
[0195]Vector Construction:
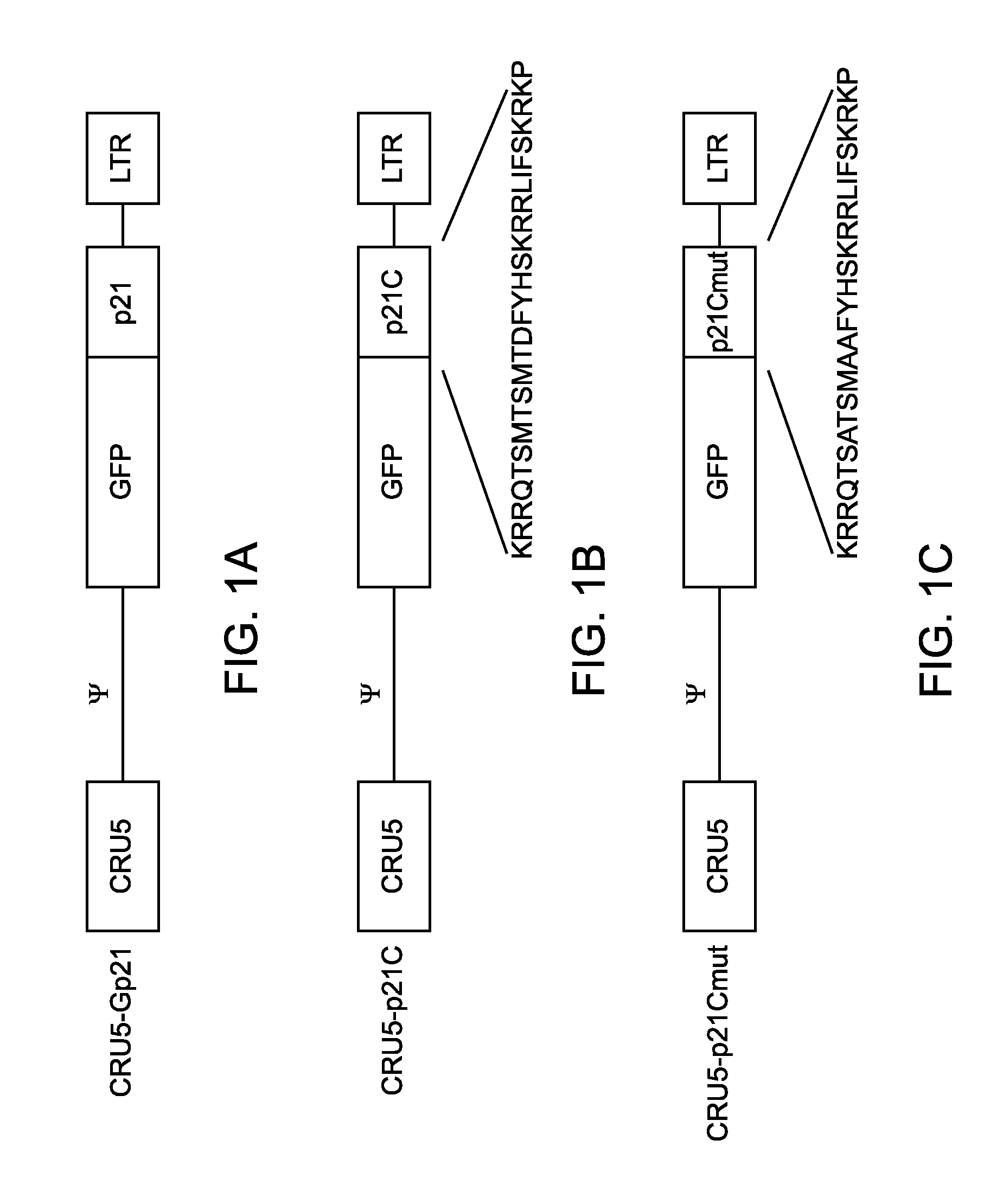
[0196]The coding region of the p21 gene was cloned from Jurkat cDNA by PCR with an upstream primer covering the start methionine (5′-GATCGGATCCACC ACCATGGGCTCAGAACCGGCTGGGGATGTC) and C-terminus (5′-GATCCC AATTTAATGGTTTTATTTGTCATCGTCATCCTTGTAGTCGGGCTTCCTCTTGGAGAAGATCAGCCG GCGTTTG). The single PCR product was directionally cloned into the CRU5-GFP retroviral vector (Rigel, Inc.) through flanking BstXI sites within the primers. The resultant construct, CRU5-GFP-p21F (FIG. 1), encodes the GFP fused (in frame) to the human p21 protein with a Gly insertion at position 2 and a FLAG-epitope at the C-terminus. The C-terminal 24 amino-acids of p21 were cloned into the CRU5-GFP retroviral vector (Rigel, Inc.) through flanking BstXI sites within the PCR primers: 5′GATCCCACCACCATGGGCAAACGGCGGCAGACCAGCATGACAGATTTCTACCACTCCAAACGCC GGCTGATCTTCTCCAA; 5′GATCCCAATTTAAATGGTTTTATTTGTCATCGTCATCCTTGTAGTC...
example 2
Population Based Exocytic Enzyme Activity Measurements
[0203]Materials:
[0204]All chemicals were obtained from Sigma Chemical Co. Dyes and glucuronide were obtained from Molecular Probes, Inc. Cell lines MC-9 and RBL-2H3 were obtained from American Type Culture Collection (ATCC). Cell culture reagents were obtained from Fisher Scientific and molecular biology reagents from Clontech Inc.
[0205]Cell Culture:
[0206]MC-9 cells were maintained as suspension cultures in flasks in media consisting of DMEM with L-arginine (116 mg / ml), L-asparagine (36 mg / ml), sodium pyruvate (1 mM), non-essential amino acids (0.1 mM), folic acid (6 mg / ml), 2-mercaptoethanol (0.05 mM), L-glutamine (2 mM), heat inactivated fetal bovine serum (10%), and 10% T-stim conditioned media (Collaborative Research, Inc.). The cells were kept at a density of between 0.25 and 2×106 / ml. Experiments were only conducted on cells which were greater than 95% viable as determined by trypan blue exclusion. RBL-2H3 cells were mainta...
example 3
Mast Cell Exocytic Light Scatter Changes
[0215]The cells were prepared as described in Example 2, and light scatter properties were determined.
[0216]Results:
[0217]The results are shown in FIG. 4. Light scatter changes observed on the flow cytometer (side scatter vs. forward scatter) are plotted as bivariate histograms for RBL-2H3 cells (A, D) and MC-9 cells (B, C, E, F). Cells were stimulated with the ionophore A23187 (0.5 ug / ml) and observed at various timepoints [0 minutes (A, C), 5 minutes (E), 10 minutes (D), and 30 minutes (B, F)]. Time dependent scatter changes are evident in both cell lines with significant changes occurring during the first 10 minutes which represents the major bolus of exocytosis in these cells.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com